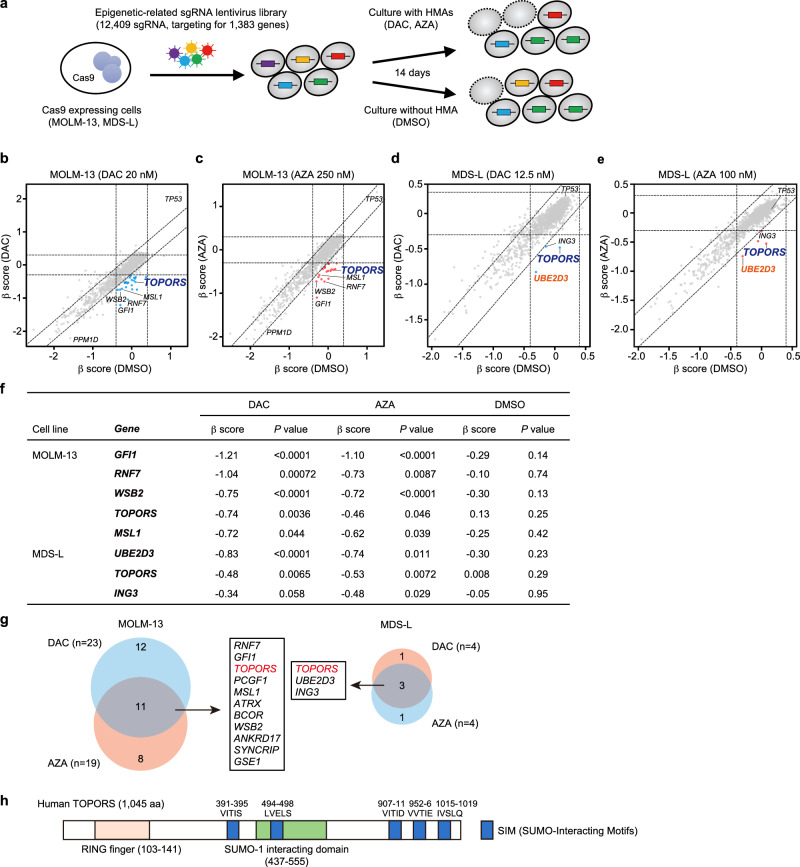
Fig. 1. CRISPR-Cas9 screening reveals that TOPORS KO augments sensitivity to HMAs.
a Outline of CRISPR-Cas9 screening with and without HMAs. Screening results using MOLM-13 cells treated with DAC at 20 nM, (b) and AZA at 250 nM, (c) and MDS-L cells treated with DAC at 12.5 nM, (d) and AZA at 100 nM (e). Scatter plots showing the β scores of each gene, which indicate changes in the sgRNA content for each gene, in the presence and absence of HMAs. Candidate genes whose sgRNAs decreased during culture specifically in the presence of HMAs are indicated by blue dots (DAC) and red dots (AZA). f List of candidate genes identified to be in common in the presence of DAC and AZA. The p-value was calculated using a two-sided test. g Venn diagrams comparing screening results and the list of candidate genes identified to be in common in the presence of DAC and AZA. (h) Schematic representation of the TOPORS protein with the RING finger domain, the SUMO-1 interacting domain, and putative SUMO-interaction motifs (SIMs). Source data are provided as a Source Data file.