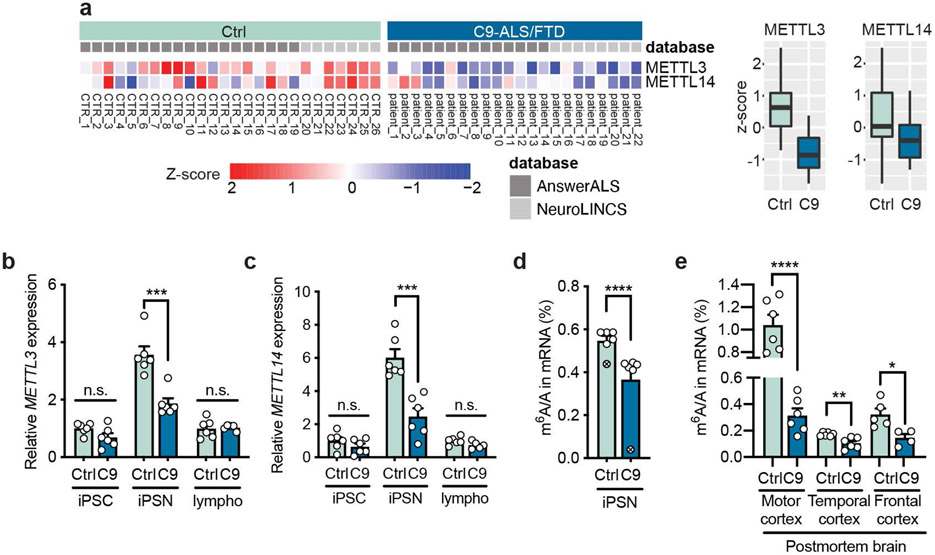
Fig. 1: The expression of m6A methyltransferases and the m6A RNA modification levels are downregulated in C9ORF72-ALS/FTD.
a, Proteomic heatmap (left) and boxplots (right) of the m6A “writer” complex core components in the iPSNs of control (n=26) and C9ORF72-ALS/FTD patients (n=22). Data were analyzed using the proteomic database from Answer ALS and NeuroLINC center. Batch controls were used for normalization of the AnswerALS data. Box plots indicate the interquartile range with the central line representing the median, and the vertical lines extend to the extreme values in the group. b,c, METTL3(b) and (c) METTL14 RNA expression levels were measured by RT-qPCR in control and C9 iPSC, iPSN and lymphoblast cells. Points represent individual cell lines, including one pair with isogenic control in which the repeat expansion was removed71. The isogenic control iPSC was set to 1. n=6 for both control and patient iPSC lines. n=6 for control and 5 for C9ORF72-ALS lymphoblast lines. Date are mean ± s.e.m. METTL3: iPSC, p=0.1042; iPSN, ***p=0.0006; lympho, p=0.8356. METTL14: iPSC, p=0.2328; iPSN, ***p=0.0006; lympho, p=0.0944. Two-tailed t test. d, UHPLC-QQQ-MS/MS quantification of the m6A/A ratio in mRNAs from iPSNs. The mRNAs of the isogenic pair (circle-cross) were extracted by poly-A selection; the mRNAs of the other iPSN lines were extracted by ribosomal RNA depletion. Two-tailed t test was performed on 5 pairs of control and C9 lines (ribosomal RNA depletion samples). Date are mean ± s.e.m. ****p<0.0001 by two-tailed t test. e, UHPLC-QQQ-MS/MS quantification of the m6A/A ratio in mRNAs from postmortem brain tissues extracted by poly-A selection. Points represent individual patient samples. n=6 for both control and patient samples. Date are mean ± s.e.m. *p=0.0168, **p=0.0039, ****p<0.0001, by two-tailed t test.