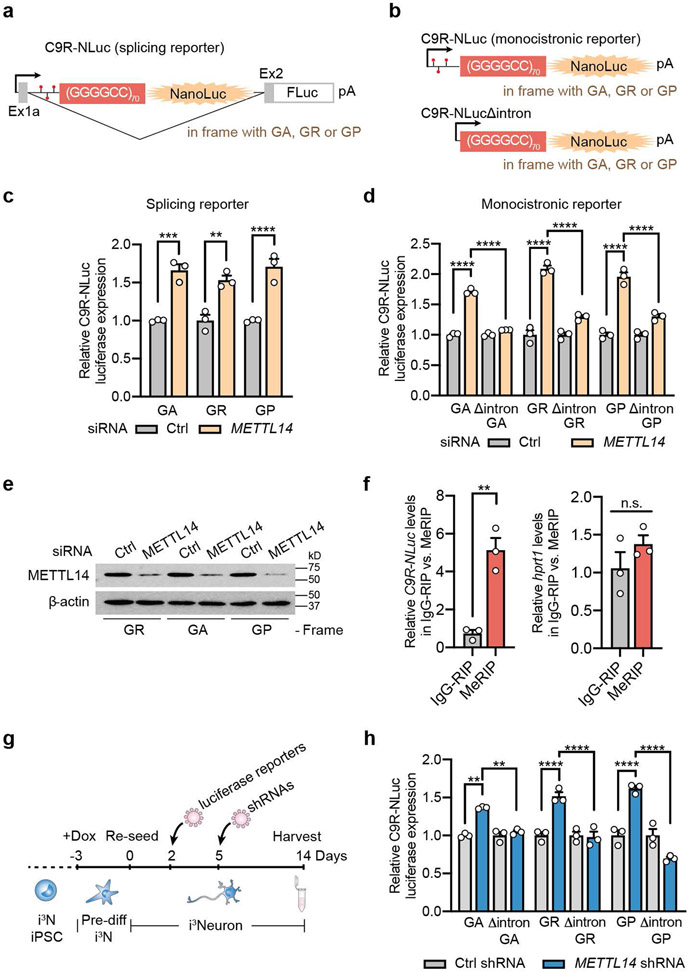
Fig. 4: The m6A modification in the repeat-containing intron modulates DPR protein production.
a, Schematic of the bicistronic splicing reporter for (GGGGCC)70 RAN translation, which mimics the splicing of C9 repeat RNA from the endogenous context. Predicted m6A sites were indicated as red lollipops in the intron sequence upstream of the repeats. b, Schematic of the monocistronic mRNA luciferase reporters for (GGGGCC)70 RAN translation with or without the intronic sequence. Predicted m6A sites were indicated as red lollipops. c, Relative DPR levels were measured by luciferase assay upon METTL14 knockdown in the HeLa Flp-In cells expressing the reporters in (a). NLuc signal was normalized to the total protein level in each sample and the relative expression was compared to non-targeting siRNA control. Data are mean ± s.e.m. GA, ***p=0.0002; GR, ** p=0.0013; GP, ****p<0.0001, by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. d, Relative DPR levels were measured by luciferase assay upon METTL14 knockdown in the HeLa Flp-In cells expressing the reporters in (b). NLuc signal was normalized to the total protein level in each sample and the relative expression was compared to non-targeting siRNA control. Data are mean ± s.e.m. ****p<0.0001, by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. e, Western blotting of METTL14 in non-targeting or METTL14 siRNA transfected HeLa Flp-In cells. β-actin was blotted as internal control. f, RNA immunoprecipitation by IgG or m6A antibody (MeRIP) followed by RT-qPCR of HPRT1 RNA (left) or C9 repeat RNA (right) in the HeLa Flp-In C9R-NLuc reporter cells. HPRT1 RNA does not have m6A modification and serves as a negative control. Data are mean ± s.e.m. Points represent three biological replicates. C9R-NLuc, **p=0.0028; HPRT1, p=0.2584, by two-tailed t test. g, Timeline for the luciferase reporter experiments in human i3Neurons. h, Relative DPR levels were measured by luciferase assay upon METTL14 knockdown in the i3Neurons expressing the reporters as described in (g). n=3 biological replicates. Data are mean ± s.e.m. **p=0.001 and 0.0053, respectively, ****p<0.0001, by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. Points represent three biological replicates in each group.