Figure 4.
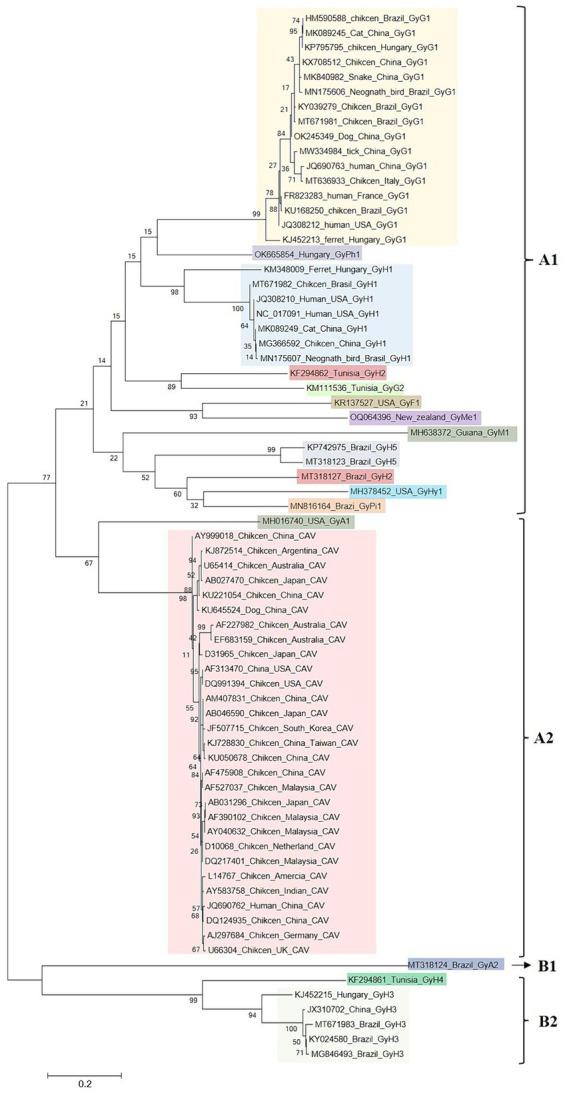
Maximum likelihood phylogenetic analyses of GyV. Duplicate sequences obtained with the same date and location were removed using BioAider with a 99% threshold. The final 71 reference sequences of GyV were collected for data analysis. All GyV sequences (3,401 bp) were aligned using Clustal W in MEGA 6. The trees were constructed with MEGA 6, using the maximum likelihood GTR + G4 as a substitution model for the genome. Phylogenetic trees Bootstrap values (with a basis on 1,000 replicates) superior to 60% are shown.
