Abstract
Background
In many cancers, specific subtypes are more prevalent in specific racial backgrounds. However, little is known about the racial distribution of specific molecular types of brain tumors. Public data repositories lack data on many brain tumor subtypes as well as diagnostic annotation using the current World Health Organization classification. A better understanding of the prevalence of brain tumors in different racial backgrounds may provide insight into tumor predisposition and development, and improve prevention.
Methods
We retrospectively analyzed the racial distribution of 1709 primary brain tumors classified by their methylation profiles using clinically validated whole genome DNA methylation. Self-reported race was obtained from medical records. Our cohort included 82% White, 10% Black, and 8% Asian patients with 74% of patients reporting their race.
Results
There was a significant difference in the racial distribution of specific types of brain tumors. Blacks were overrepresented in pituitary adenomas (35%, P < .001), with the largest proportion of FSH/LH subtype. Whites were underrepresented at 47% of all pituitary adenoma patients (P < .001). Glioblastoma (GBM) IDH wild-type showed an enrichment of Whites, at 90% (P < .001), and a significantly smaller percentage of Blacks, at 3% (P < .001).
Conclusions
Molecularly classified brain tumor groups and subgroups show different distributions among the three main racial backgrounds suggesting the contribution of race to brain tumor development.
Keywords: CNS tumor prevalence, DNA methylation, ethnicity, race
Key Points.
Current epidemiological studies of brain tumors do not account for molecularly defined classification.
Using DNA methylation and self-reported race, we show that different types of brain tumors have different prevalence in different racial backgrounds.
Our data suggest the contribution of race to brain tumor development.
Importance of the Study.
Association between race and specific types of cancer is well established across multiple tumor types. However, in brain tumors, the association between molecularly defined brain tumor types and race is not well understood. Current epidemiological registries such as SEER, CBTRUS, and NPCR lack comprehensive records for multiple tumor types and races, and are for the majority of tumor types annotated using histopathology not reflecting recent molecular classification and molecular subtypes. Using DNA methylation as a clinical diagnostic standard, and self-reported race in medical records, we show that specific molecularly defined brain tumor types are more frequent in specific racial backgrounds. Whites showed enrichment for IDH mutant diffuse gliomas and IDH wild-type Glioblastoma, while Blacks showed significantly decreased prevalence of GBM IDH wild-type, but significant enrichment for pituitary adenoma. In addition, our study also highlights shortcomings of self-reported race and the need for concurrent genotyping analysis and molecular brain tumor analyses.
Although central nervous system (CNS) tumors represent only 1% of newly diagnosed tumors in the United States, they are the 10th leading cause of death in adults, the most common solid and malignant tumors in children under 15 years of age, and the leading cause of cancer-associated death in pediatric patients and young adults.1–3 Recent advances in molecular profiling have identified multiple molecularly distinct subgroups of primary CNS tumors, which vary in their molecular drivers, biological behavior, and clinical outcomes. For example, diffuse gliomas have been completely reclassified using a combination of IDH1/2, TERT promoter, ATRX, and TP53 mutational status and chromosomal status of chromosomal arms 1p and 19q.4
Associations between race and certain cancers, including molecular subgroups, have been identified in multiple cancer types. For example, in non-small cell lung cancer, EGFR mutations are more common in young Asian females, while in melanoma, BRAF and NRAS mutations are more common in White patients, and triple-negative breast cancer (ER-, PR-, and HER2-) is more prevalent in Black women.5 Molecular genetic information can provide insight in the genetic underlying of cancer and specific risks for community-based prevention.
Currently, the most comprehensive epidemiological collection of data relating brain tumor type to race is the National Cancer Institute’s (NCI) Surveillance, Epidemiology, and End Results Program (SEER). However, the SEER registry lacks molecular characterization as well as comprehensive records across races and years. The World Health Organization (WHO) classification of CNS tumors now recognizes more than 100 molecularly distinct tumor entities. However, the SEER database only includes the larger, nonspecific, and currently outdated entities such as Diffuse Astrocytoma and Anaplastic Astrocytoma, Glioblastoma, Other Glioma, Embryonal Tumors, Meningioma, and Other Tumors lacking critical molecular information. Other databases such as the Central Brain Tumor Registry of the United States (CBTRUS), the Center for Disease Control’s (CDC) database, and the National Program of Cancer Registries (NPCR) exhibit similar shortcomings.6 While the most recent CBTRUS report6 includes some molecular characterization, such as IDH status for diffuse gliomas and molecular subgroups of medulloblastoma, it is limited both in terms of molecular subtypes and reported races. Most notably, it is limited to adult-type diffuse glioma, IDH mutant, Glioblastoma IDH wild-type, medulloblastoma, and some rare tumors such as K27M mutated glioma, ETMR, and ependymoma RELA. Furthermore, it only includes incidence in White, Black, and Hispanic / Non-Hispanic patients, while lacking any molecularly annotated data for Asian patients. Race and ethnicity data are also lacking for the Ependymoma RELA category and no data are available for Black patients for most medulloblastoma subtypes or ETMR C19MC altered. This highlights the need for brain tumor research with both comprehensive molecular analysis and race annotation.
Whole genome DNA methylation analysis has emerged in recent years as an accurate pan-CNS tumor molecular classification method to distinguish between more than a hundred molecularly defined subgroups of CNS tumors. DNA methylation-based classification enables diagnostic standardization between laboratories reducing diagnostic errors and variability between laboratories.7–9
The 2021 edition of the WHO classification of CNS tumors has incorporated the use of DNA methylation profiling and the majority of WHO tumor entities have a distinct methylation signature and providing a standardized molecular classification framework across all brain tumor subgroups.10 Therefore, epidemiological CNS studies should incorporate molecularly classification to properly asses the distribution of different brain tumor types across racial subgroups.
In this study, we aimed to delineate predisposition to molecularly defined primary CNS tumors across racial groups utilizing DNA methylation-based molecular and self-reported race of 1709 patients.
Methods
Cohort Criteria
We retrospectively analyzed data of 1709 primary CNS tumors diagnosed and operated on at NYU Langone Health (NYULH) between 2015 and 2022. Race and DNA methylation results were retrieved from medical records. A complete list of diagnoses and number of patients for each category is available in Table 1, and a list of abbreviations and complete names for each entity is in Supplementary Table 1. A detailed description of each DNA methylation class is available on www.molecularneuropathology.org. Outside cases profiled in consultation lacked ethnicity data and were therefore excluded from the study.
Table 1.
Molecular Groups, Subgroups Defined by Methylation, and Race Distribution. Bolded rows represent molecular groups. A Complete List With Abbreviations and Full Names for Each Entity is in Supplementary Material
| Methylation Class | White | Asian | Black | More than one | Other | Unknown | Total |
|---|---|---|---|---|---|---|---|
| EPN | 54 (45.8%) | 10 (8.5%) | 5 (4.2%) | 1 (0.8%) | 16 (13.6%) | 32 (27.1%) | 118 |
| EPN_MPE | 10 | 2 | 0 | 0 | 3 | 1 | 16 |
| EPN_PF_A | 11 | 0 | 2 | 0 | 5 | 6 | 24 |
| EPN_PF_B | 5 | 3 | 2 | 0 | 0 | 7 | 17 |
| EPN_RELA | 10 | 3 | 1 | 1 | 1 | 7 | 23 |
| EPN_SPINE | 6 | 2 | 0 | 0 | 5 | 3 | 16 |
| EPN_YAP | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| SUBEPN_PF | 10 | 0 | 0 | 0 | 2 | 7 | 19 |
| SUBEPN_ST | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| LGG | 91 (51.1%) | 6 (3.4%) | 19 (10.7%) | 1 (0.5%) | 34 (19.1%) | 27 (15.2%) | 178 |
| DLGNT | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| LGG_DIG_DIA | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| LGG_DNT | 7 | 0 | 0 | 0 | 4 | 2 | 13 |
| LGG_GG | 6 | 0 | 1 | 0 | 2 | 4 | 13 |
| LGG_MYB | 5 | 1 | 1 | 0 | 4 | 1 | 12 |
| LGG_RGNT | 3 | 1 | 1 | 0 | 0 | 0 | 5 |
| LGG_PA_GG_ST | 11 | 0 | 2 | 1 | 5 | 6 | 25 |
| LGG_PA_MID | 15 | 0 | 2 | 0 | 3 | 2 | 22 |
| LGG_PA_PF | 21 | 2 | 9 | 0 | 11 | 8 | 51 |
| LGG_SEGA | 4 | 0 | 2 | 0 | 1 | 4 | 11 |
| PXA | 17 | 2 | 1 | 0 | 4 | 0 | 24 |
| MB | 99 (67.3%) | 2 (1.4%) | 6 (4.1%) | 0 (0.0%) | 16 (10.9%) | 24 (16.3%) | 147 |
| MB_WNT | 10 | 0 | 1 | 0 | 0 | 1 | 12 |
| MB_G3 | 21 | 0 | 2 | 0 | 0 | 6 | 29 |
| MB_G4 | 42 | 2 | 0 | 0 | 4 | 4 | 52 |
| MB_SHH_CHL_AD | 12 | 0 | 3 | 0 | 5 | 12 | 32 |
| MB_SHH_INF | 14 | 0 | 0 | 0 | 7 | 1 | 22 |
| PIN | 12 (57.1%) | 2 (9.5%) | 1 (4.8%) | 0 (0.0%) | 1 (4.8%) | 5 (2.4%) | 21 |
| PIN_T_PB_B | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PIN_T_PB_A | 6 | 0 | 0 | 0 | 0 | 0 | 6 |
| PIN_T_PPT | 2 | 2 | 1 | 0 | 1 | 2 | 8 |
| PTPR_A | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| PTPR_B | 3 | 0 | 0 | 0 | 0 | 3 | 6 |
| PITAD | 27 (38.0%) | 10 (14.1%) | 20 (28.2%) | 0 (0.0%) | 11 (15.5%) | 4 (5.6%) | 72 |
| PITAD_ACTH | 6 | 2 | 2 | 0 | 4 | 2 | 16 |
| PITAD_FSH_LH | 13 | 5 | 14 | 0 | 4 | 2 | 38 |
| PITAD_STH_DNS_A | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| PITAD_STH_DNS_B | 4 | 2 | 1 | 0 | 1 | 0 | 8 |
| PITAD_STH_SPA | 2 | 0 | 2 | 0 | 1 | 0 | 5 |
| PITAD_TSH | 1 | 1 | 1 | 0 | 1 | 0 | 4 |
| MNG | 179 (63.7%) | 23 (8.2%) | 31 (11.0%) | 0 (0.0%) | 23 (8.2%) | 25 (8.9%) | 281 |
| BEN-1 | 45 | 3 | 6 | 0 | 11 | 6 | 71 |
| BEN-2 | 57 | 10 | 8 | 0 | 4 | 4 | 83 |
| BEN-3 | 28 | 4 | 11 | 0 | 5 | 7 | 55 |
| INT-A | 41 | 5 | 6 | 0 | 2 | 4 | 58 |
| INT-B | 5 | 1 | 0 | 0 | 0 | 1 | 7 |
| MAL | 3 | 0 | 0 | 0 | 1 | 2 | 6 |
| SMARCE | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| MTGF_GBM | 253 (69.9%) | 19 (5.2%) | 9 (2.5%) | 1 (0.2%) | 41 (11.4%) | 39 (10.8%) | 362 |
| GBM_MES | 57 | 4 | 3 | 0 | 10 | 11 | 85 |
| GBM_MID | 18 | 6 | 1 | 0 | 1 | 2 | 28 |
| GBM_MYCN | 4 | 1 | 0 | 0 | 0 | 5 | 10 |
| GBM_RTK_I | 73 | 2 | 0 | 0 | 10 | 7 | 92 |
| GBM_RTK_II | 99 | 3 | 4 | 0 | 19 | 14 | 139 |
| GBM_RTK_III | 2 | 3 | 0 | 0 | 1 | 0 | 6 |
| MTGF_IDH_GLM | 136 (63.8%) | 12 (5.9%) | 5 (2.4%) | 1 (0.5%) | 37 (17.4%) | 22 (10.3%) | 213 |
| O_IDH | 38 | 4 | 1 | 1 | 16 | 6 | 66 |
| A_IDH | 57 | 3 | 2 | 0 | 13 | 10 | 85 |
| A_IDH_HG | 41 | 5 | 2 | 0 | 8 | 6 | 62 |
| SCHW | 38 (65.5%) | 6 (10.3%) | 3 (5.2%) | 0 (0.0%) | 8 (13.8%) | 3 (5.2%) | 58 |
| DMG_K27 | 29 | 2 | 6 | 1 | 6 | 6 | 50 |
Race Self-identification
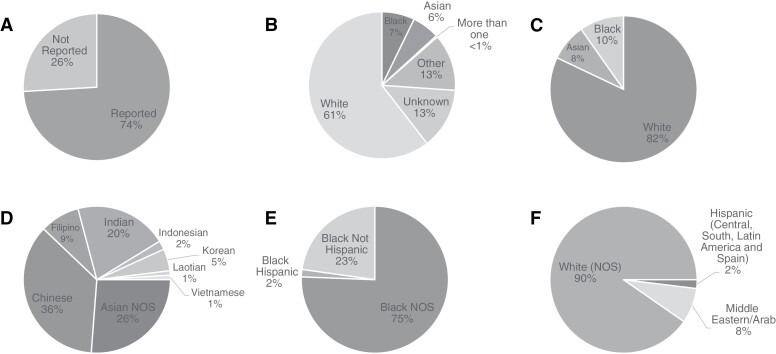
For each patient, we retrieved self-reported race/ethnicity from electronic medical records. Self-reported race/ethnicity is collected at NYULH when patients first register at the hospital and are independent of this study. Declaration of race/ethnicity is not required, and the patient could choose not to respond (Figure 1A) and was placed into the “Unknown” category. If the patient did not identify as any one race/ethnicity, they could respond as “More than one” and if the patient did not feel represented by any race/ethnicity, they could respond as “Other” (Figure 1B).
Figure 1.
Overall cohort racial demographics. This figure shows the racial breakdown of our cohort, overall, and within each group. (A) Proportion of cohort that reported race or ethnicity. Patients who reported “Other” or “Unknown” are assigned to the “Not Reported” category (n = 1709). (B) Composition of our total cohort, both those who did and did not report race. This includes More than One, Other, and Unknown (n = 1709). (C) The racial breakdown of the final cohort stratified into Asian, Black, and White (n = 1260). (D) Total distribution of ethnicity within Asian (n = 103). (E) Total distribution of ethnicity within Black (n = 123). F- Total distribution of ethnicity within White (n = 1034).
Collected responses varied widely in levels of specificity, from broad descriptors such as “Asian,” “Black,” or “White,” to specific countries of origin such as “Laotian” or “Honduran.” In these cases, we followed the NIH guidelines on Race and National Origin to group these respondents into larger groups as Asian, Black, or White (Figure 1C).11 Patients who did not fit into one of these categories were grouped into “Other” category (Supplementary Figure 1).
Patients who self-reported as “Hispanic” may fall into any of the three main groups, White, Black, or Asian, and those who did not provide any other specification were placed into “Other.” No patients self-identified as both “Hispanic” and “Asian” (Figure 1D-F).
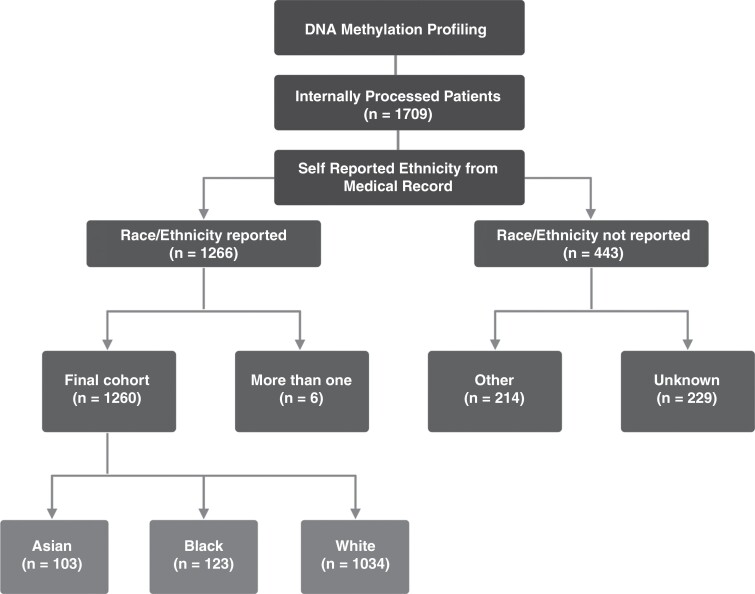
Patients were grouped into Asian, Black, and White to best represent the diversity of our cohort while maintaining group sizes large enough to perform statistical analyses (Figure 2). These classifications were created in accordance with the NIH guidelines on Race and National Origin.11 Patients with no race information were excluded from further analysis. We do not have any information on whether some patients would be more likely to not self-report race. The cohort design and analysis workflow is shown in Figure 3.
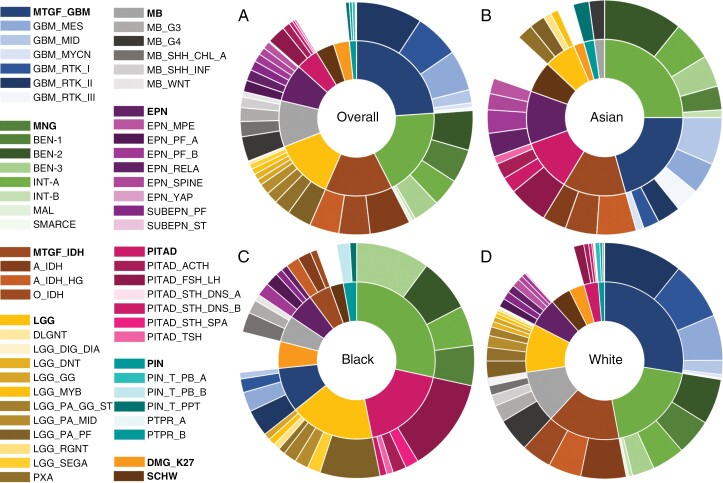
Figure 2.
Breakdown of brain tumor types within each racial group. This figure shows the distribution of brain tumor groups and their subgroups in each racial group. (A) Distribution of brain tumor groups and subgroups overall (Asian, Black, and White) (B) Distribution of brain tumor groups and subgroups in Asian. (C) Distribution of brain tumor groups and subgroups in Black. (D) Distribution of brain tumor groups and subgroups in White.
Figure 3.
Study design and selection criteria.
DNA Methylation Tumor Classification
Molecular profiling of brain tumors was performed using clinically validated NY State-approved genome-wide DNA methylation profiling12 and classified as described previously,7 along with results retrieved from electronic medical records. A complete list of DNA methylation classes is available in Supplementary Table 1.
Statistical Analysis
Our study was designed to explore potential differences in ethnic distribution across various molecular types and subtypes of brain tumors. We hypothesized that certain ethnicities could demonstrate statistically significant over- or underrepresentation within particular types/subtypes. To rigorously examine these hypotheses, we utilized Fisher’s Exact Test, a robust statistical method devised to detect nonrandom associations between two categorical variables, making it suitable for studies with small sample sizes.13
In addition, to account for the potential inflation in Type I errors due to the multiple tests and comparisons performed, we also employed the Holm-Bonferroni Correction method.14 This method arranges all P-values from the tests in ascending order and compares them with different thresholds (ie, adjusted significance level) to determine whether it is statistically significant. The smallest P-value is compared with the threshold α/k, the next with α/(k-1), where α is our original significance level of 0.05, and k is the total number of tests. We considered a P-value significant if it was less than its respective threshold. Once a P-value was found exceeding its adjusted significance level, all subsequent p-values were treated as nonsignificant.
The racial composition of our cohort encompassed 82% White, 10% Black, and 8% Asian participants. For each tumor type (such as low-grade gliomas), we first tested whether the ethnic distribution within this type was the same as that of the overall tumor cohort. Upon finding differences, we proceeded to test whether the ethnic distributions of one ethnicity versus the remaining ethnicities (eg, White vs. nonwhite, Asian vs. non-Asian, Black vs. non-Black) within the specific tumor type mirror those in the whole cohort. A rejection of the null hypothesis using the Holm-Bonferroni adjusted significance level showed notable over- or underrepresentation of the ethnicity under consideration, compared to the remaining ethnicities.
Our subsequent analytical stage involved testing whether the ethnic distribution for a particular subtype (eg, receptor tyrosine kinase III glioblastomas) is the same as that of the parent tumor type (eg, glioblastomas). If discrepancies were detected, we followed a similar procedure as before, testing whether the ethnic distributions of one ethnicity versus the remaining ethnicities within the specific tumor subtype mirror those in the parent tumor type. This also constituted a one-sided test, with a rejection of the null hypothesis using the Holm-Bonferroni adjusted significance level indicating a significant over- or underrepresentation of the ethnicity in consideration relative to the remaining ethnicities. See Table 2 for a comprehensive description of the results.
Table 2.
Statistical Analyses of the Molecularly Defined Tumor Types and Racial Distribution
| Test | Group | Subgroup | P value | Statistic White | P value White | Statistic Asian | P value Asian | Statistic Black | P value Black |
|---|---|---|---|---|---|---|---|---|---|
| 1 | EPN | NA | .241 | 0.739 | .190 | 1.859 | .069 | 0.869 | .475 |
| 2 | EPN | EPN_MPE | .737 | 1.475 | .481 | 1.197 | .558 | 0.000 | .585 |
| 3 | EPN | EPN_PF_A | .301 | 1.621 | .427 | 0.000 | .350 | 1.921 | .366 |
| 4 | EPN | EPN_PF_B | .132 | 0.302 | .079 | 2.534 | .203 | 2.624 | .261 |
| 5 | EPN | EPN_RELA | .872 | 0.744 | .438 | 1.626 | .371 | 0.822 | .670 |
| 6 | EPN | EPN_SPINE | .649 | 0.890 | .594 | 1.979 | .356 | 0.000 | 1.000 |
| 7 | EPN | EPN_YAP | 1.000 | Inf | .603 | 0.000 | 1.000 | 0.000 | 1.000 |
| 8 | EPN | SUBEPN_PF | .499 | 2.932 | .274 | 0.000 | .343 | 1.066 | .655 |
| 9 | EPN | SUBEPN_ST | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 |
| 10 | LGG | NA | .069 | 0.797 | .203 | 0.609 | .165 | 1.814 | .024 |
| 11 | LGG | DLGNT | 1.000 | Inf | .786 | 0.000 | 1.000 | 0.000 | 1.000 |
| 12 | LGG | LGG_DIG_DIA | 1.000 | Inf | .786 | 0.000 | 1.000 | 0.000 | 1.000 |
| 13 | LGG | LGG_DNT | .718 | Inf | .195 | 0.000 | 1.000 | 0.000 | .594 |
| 14 | LGG | LGG_GG | 1.000 | 1.642 | .543 | 0.000 | 1.000 | 0.852 | .682 |
| 15 | LGG | LGG_MYB | .408 | 0.689 | .480 | 3.013 | .344 | 0.852 | .682 |
| 16 | LGG | LGG_RGNT | .182 | 0.416 | .310 | 4.479 | .262 | 1.274 | .602 |
| 17 | LGG | LGG_PA_GG_ST | 1.000 | 1.507 | .460 | 0.000 | 1.000 | 0.929 | .644 |
| 18 | LGG | LGG_PA_MID | .886 | 2.051 | .281 | 0.000 | 1.000 | 0.682 | .473 |
| 19 | LGG | LGG_PA_PF | .260 | 0.527 | .105 | 1.220 | .548 | 1.987 | .108 |
| 20 | LGG | LGG_SEGA | .484 | 0.553 | .398 | 0.000 | 1.000 | 2.528 | .275 |
| 21 | LGG | PXA | .282 | 1.552 | .371 | 2.024 | .334 | 0.271 | .163 |
| 22 | MB | NA | .010 | 2.708 | .002 | 0.213 | .008 | 0.551 | .105 |
| 23 | MB | MB_WNT | .599 | 0.810 | .599 | 0.000 | 1.000 | 1.674 | .505 |
| 24 | MB | MB_G3 | .752 | 0.850 | .559 | 0.000 | 1.000 | 1.597 | .430 |
| 25 | MB | MB_G4 | .192 | 1.692 | .400 | 2.483 | .332 | 0.000 | .181 |
| 26 | MB | MB_SHH_CHL_AD | .140 | 0.327 | .135 | 0.000 | 1.000 | 4.134 | .081 |
| 27 | MB | MB_SHH_INF | 1.000 | Inf | .362 | 0.000 | 1.000 | 0.000 | 1.000 |
| 28 | PIN | NA | .260 | 0.526 | .180 | 1.488 | .417 | 1.984 | .231 |
| 29 | PIN | PIN_T_PB_B | .123 | 0.000 | .123 | 0.000 | 1.000 | Inf | .058 |
| 30 | PIN | PIN_T_PB_A | .745 | Inf | .184 | 0.000 | 1.000 | 0.000 | .539 |
| 31 | PIN | PIN_T_PPT | .475 | 0.296 | .233 | 4.544 | .210 | 1.158 | .675 |
| 32 | PIN | PTPR_A | 1.000 | Inf | .722 | 0.000 | 1.000 | 0.000 | 1.000 |
| 33 | PIN | PTPR_B | 1.000 | Inf | .399 | 0.000 | 1.000 | 0.000 | 1.000 |
| 34 | PITAD | NA | 1.100E-08* | 0.197 | 1.120E-08* | 2.373 | .021 | 4.997 | 5.680E-07* |
| 35 | PITAD | PITAD_ACTH | .657 | 1.654 | .347 | 1.172 | .574 | 0.467 | .292 |
| 36 | PITAD | PITAD_FSH_LH | .733 | 0.763 | .348 | 0.872 | .532 | 1.433 | .280 |
| 37 | PITAD | PITAD_STH_DNS_A | 1.000 | Inf | .483 | 0.000 | 1.000 | 0.000 | 1.000 |
| 38 | PITAD | PITAD_STH_DNS_B | .657 | 1.472 | .464 | 1.859 | .391 | 0.313 | .258 |
| 39 | PITAD | PITAD_STH_SPA | 1.000 | 1.109 | .655 | 0.000 | 1.000 | 1.830 | .457 |
| 40 | PITAD | PITAD_TSH | .768 | 0.561 | .551 | 2.309 | .462 | 0.926 | .722 |
| 41 | MNG | NA | .153 | 0.726 | .041 | 1.222 | .241 | 1.422 | .069 |
| 42 | MNG | BEN-1 | .571 | 1.506 | .197 | 0.538 | .239 | 0.815 | .431 |
| 43 | MNG | BEN-2 | .614 | 0.955 | .498 | 1.403 | .259 | 0.779 | .354 |
| 44 | MNG | BEN-3 | .134 | 0.564 | .078 | 0.937 | .585 | 2.232 | .039 |
| 45 | MNG | INT-A | .964 | 1.124 | .456 | 0.971 | .596 | 0.850 | .469 |
| 46 | MNG | INT-B | .583 | 1.506 | .581 | 1.820 | .474 | 0.000 | 1.000 |
| 47 | MNG | MAL | 1.000 | Inf | .457 | 0.000 | 1.000 | 0.000 | 1.000 |
| 48 | MNG | SMARCE | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 |
| 49 | MTGF_GBM | NA | .001* | 1.909 | .001* | 0.807 | .245 | 0.341 | 2.521E-04* |
| 50 | MTGF_GBM | GBM_MES | .829 | 0.934 | .513 | 0.923 | .574 | 1.336 | .443 |
| 51 | MTGF_GBM | GBM_MID | .015 | 0.296 | .017 | 4.337 | .010 | 1.133 | .613 |
| 52 | MTGF_GBM | GBM_MYCN | .427 | 0.460 | .427 | 3.436 | .305 | 0.000 | 1.000 |
| 53 | MTGF_GBM | GBM_RTK_I | .396 | 2.088 | .123 | 0.370 | .133 | 0.726 | .506 |
| 54 | MTGF_GBM | GBM_RTK_II | .353 | 1.619 | .181 | 0.404 | .104 | 1.066 | .561 |
| 55 | MTGF_GBM | GBM_RTK_III | .006* | 0.078 | .011 | 20.187 | .004 | 0.000 | 1.000 |
| 56 | MTGF_IDH | NA | .019 | 1.751 | .020 | 0.950 | .512 | 0.313 | .003 |
| 57 | MTGF_IDH | O_IDH | .906 | 0.950 | .555 | 1.204 | .482 | 0.706 | .607 |
| 58 | MTGF_IDH | A_IDH | .858 | 1.423 | .347 | 0.599 | .324 | 0.987 | .675 |
| 59 | MTGF_IDH | A_IDH_HG | .656 | 0.733 | .337 | 1.364 | .381 | 1.285 | .530 |
| 60 | SCHW | NA | .438 | 0.925 | .479 | 1.633 | .196 | 0.632 | .322 |
| 61 | DMG_K27 | NA | .415 | 0.794 | .348 | 0.638 | .408 | 1.792 | .154 |
*Indicates significance after Holm-Bonferroni correction
All analyses were executed using R software, version 4.0 (R Foundation for Statistical Computing).
This study was performed in accordance with the approval of the NYU Langone Institutional Review Board (IRB) and in accordance with its policy and guidelines, IRB#: i14-00948.
Results
Our cohort included 1709 patients in total, with 1034 (61%) patients in the White category, 123 (7%) patients in the Black category, and 103 (6%) patients in the Asian category, with the remaining patients in the Other (13%), More than One (<1%), and Unknown (13%) categories (Figure 1B). Out of all of our patients, 1,266 reported their race/ethnicity, and 443 patients in the categories of Other and Unknown did not report their race/ethnicity (Figure 1A). There were 6 patients in the More than One group. The remainder cohort included 1,260 patients in the groups of Asian (n = 103, 8.2%), Black (n = 123, 9.8%), and White (n = 1034, 82%; Figure 1C). There was additional diversity within all three groups (Figure 1D-F). Age and sex distribution for each molecular subtype in our cohort are included in Supplementary Table 2.
Using DNA methylation, our final cohort was classified into methylation groups with tumors further subclassified into relevant molecular subgroups as described previously7 (Figure 2A).
Glioblastoma, IDH Wild-Type
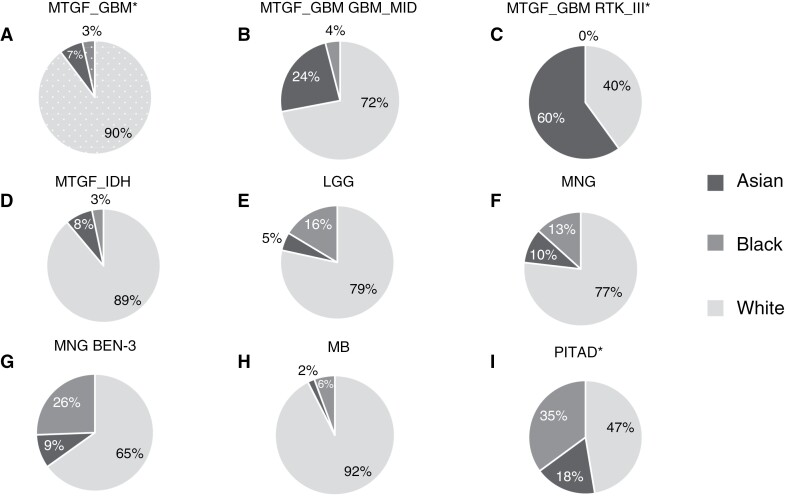
Recent WHO classification requires a lack of IDH1/2 gene mutation as a defining feature of Glioblastoma.4 Glioblastomas represented 21.2% of our total cohort, with 362 cases (Figure 2A). However, among White patients, GBM represented almost 25% of brain tumors, while it constituted only 8% of all CNS tumors in the Black group, and approximately 18% of all CNS tumors in the Asian group (Figure 2B-D). GBM IDH wild-type was more prevalent among White patients (P < .001), and less prevalent among Black patients (P < .001), with these results remaining significant after the Holm-Bonferroni correction (Figure 4A). Interestingly, the Asian patient group showed a higher prevalence of two molecularly defined GBM subgroups, Midline (P = .01) and RTK III (P = .004; Figure 4B, C). Conjunctly, the White group was underrepresented in both Midline (P = .017) and RTK III (P = .011) while Black was not. The difference in the racial composition of the glioblastoma family is different from that of the total cohort (P < .001), as are compositions of both subgroups from the GBM distribution (P = .01 and P = .006), the former of which remains significant after Holm-Bonferroni correction.
Figure 4.
Racial distribution in major methylation groups and subgroups. This figure shows the distribution of brain tumor groups and their subgroups in each racial group. Patterned background and asterisk indicate statistical significance in the racial group after Holm-Bonferroni adjustment for multiple tests, see also Table 2. (A) Distribution of glioblastoma methylation group (MTGF_GBM; n = 282). The racial distribution of the GBM family remained significantly different after adjustment from the overall racial distribution (P < .001). The proportion of the White group was significantly increased (P < .001), and the proportion of the Black group was significantly decreased (P < .001). (B) Distribution of midline glioblastoma methylation subgroup (GBM_MID; n = 25). (C) Distribution of RTK III methylation subgroup (RTK_III; n = 5). The racial composition of this methylation subgroup remained significantly different compared to the composition of the GBM group after adjustment (P = .006). (D) Distribution of IDH mutant glioma methylation group (MTGF_IDH; n = 153). (E) Distribution of low-grade glioma methylation group (LGG; n = 116). F. Distribution of meningioma methylation group (MNG; n = 233). (G) Distribution of meningioma benign class 3 methylation subgroup (BEN-3; n = 43). (H) Distribution of medulloblastoma methylation group (MB; n = 107). (I) Distribution of pituitary adenoma methylation group (PITAD; n = 57). The racial distribution of the PITAD family remained significantly different from the overall racial distribution after adjustment (P < .001). The White group was significantly underrepresented (P < .001), and the black group was significantly overrepresented (P < .001).
Other groups that were not statistically significant but showed overrepresentation of the White group included the RTKI and RTKII subgroups (Supplementary Figure 2A, B).
IDH Glioma
IDH mutant diffuse gliomas have significantly better survival than IDH wild-type diffuse gliomas (GBM) and are defined by the presence of mutations in IDH1 or IDH2 genes.4 Our cohort included 213 (12.5%) cases of IDH mutant gliomas across all three racial groups, (Figure 2A). In the White group, IDH mutant tumors represented 13.2%, in the Asian group 11.7%, and in the Black group 4.07% of all CNS tumors (Figure 2B-D). The overall racial composition of the IDH mutant glioma cohort was different from the overall cohort racial distribution (P = .02).
IDH mutant gliomas showed an overrepresentation of the White group at 89% of all IDH mutant gliomas (P = .02) and an underrepresentation of the Black group at only 3% of cases (P = .003), but neither for the Asian group (Figure 4D).
IDH mutant glioma subgroups 1p/19q co-deleted oligodendroglioma (O_IDH), astrocytoma (A_IDH), and high-grade astrocytoma (A_IDH_HG) showed similar distribution suggesting that all IDH mutant glioma subgroups have a higher prevalence in White patients and lower in Black patients (Supplementary Figure 2C-E).
Low-Grade Glioma
Low-grade gliomas (LG) are the most common primary CNS tumors in children. Our cohort included 188 tumors (11.0% of the total cohort; Figure 2A), with 8.8% of the White group, 15.4% of the Black group, and 5.83% of tumors of the Asian group of all CNS tumors (Figure 2B-D). The methylation group of low-grade gliomas showed an overrepresentation in the Black group (P = .0236; Figure 4E). Interestingly, not all LGG subgroups showed increased prevalence among Black patients and the increased prevalence seems to be due to the posterior fossa pilocytic astrocytoma subgroup. Of the patients in the posterior fossa pilocytic astrocytoma subgroup (LGG_PA_PF) Black patients represented 28% of all cases, and White patients only 66% of cases, while in the Pleomorphic Xanthoastrocytoma subgroup and Supratentorial Pilocytic Astrocytoma / Ganglioglioma subgroup (LGG_PA_GG_ST) Black group represented only 5% and 15% respectively, and White group represented 85% in both subgroups (Supplementary Figure 2F and G). Albeit not significant due to small numbers, it suggests that even within a category of LGG group, there may be further variability among different racial groups.
Meningioma
Previous studies have identified specific molecular subgroups of meningiomas associated with prognosis.15 Our cohort included 281 tumors, comprising 16.4% of total tumors (Figure 2A). Meningiomas represented 17.3% of the White group, 25.2% of the Black group, and 22.3% of the Asian group of all CNS tumors (Figure 2B-D). Meningioma showed a difference in racial composition across all meningiomas, as well as one DNA methylation subgroup, Benign 3. There was a decreased proportion of the White group at 77% (P = .04) among meningioma patients (Figure 4F). However, in the Benign 3 subgroup, Black patients made up double the percentage that they showed in the overall family, with an increase from 13% to 26% (P = .04), as the White group showed an even more pronounced decrease (65% of cases; Figure 4G).
Medulloblastoma
Medulloblastoma (MB) is the most common malignant brain tumor in children and multiple studies have highlighted the importance of molecular subclassification using DNA methylation analyses.16–18 Current CBTRUS molecular data are limited to Shh TP53 mutant and wild-type, Wnt-activated, and non-Wnt / non-Shh subtype. There is no separation of Group 3 and Group 4, and there are no molecular data for Asian patients with medulloblastoma and no data for Black patients for Wnt and non-Wnt/non-Shh categories.6 We identified 147 (8.6%) tumors in our total cohort (Figure 2A). There was a difference in the overall distribution of racial groups in Medulloblastomas compared to the total CNS tumor cohort (P = .01; Figure 4H). In the White group, MB represented 9.57%, 4.88% in the Black group, and only 1.94% of cases in the Asian group of all CNS tumors (Figure 2B-D). Medulloblastomas showed a higher representation of the White group at 92% (P = .002) and a decreased percentage of the Asian group at 2% of MB cases (P = .008; Figure 4H).
While each of the molecular MB subgroups had too few samples to reach statistical significance, the subgroup SHH A child and adult showed a difference in overall racial distribution when compared to the overall Medulloblastoma family distribution an overrepresentation of the Black group at 20% (Supplementary Figure 2H).
Pituitary Adenoma
Our cohort included 72 pituitary adenomas (4.21% of the total cohort), with 2.61% of the White group, 16.0% of the Black group, and 9.71% of the Asian group of all CNS tumors (Figure 2A-D). Pituitary adenomas showed a marked difference in racial composition compared to the total cohort (P < .001). Specifically, pituitary adenomas showed a large proportion among Black and Asian patients at 35% (P < .001) and 18% (P = .02), respectively. In contrast, there was a decreased proportion of White patients at 47% (P < .001; Figure 4I). These results remained statistically significant following the Holm-Bonferroni correction. While the number of cases was too small to reach statistical significance for molecular subgroups, the Black group was overrepresented in the Follicular Stimulating Hormone/Luteinizing Hormone (PITAD_FSH_LH, gonadotroph) subgroup (44% of cases) and the Asian group was overrepresented in the Adrenocorticotropic hormone (PITAD_ACTH) subgroup (20% of cases; Supplementary Figure 2I, J).
Other CNS Tumors
We did not identify statistically significant results concerning the remainder of families or subgroups largely due to a low number of cases and high number of tumor entities and the rarity of multiple CNS tumor types. Nevertheless, some molecular groups and subgroups showed interesting trends that ought to be investigated in larger studies. Notably, ependymomas were composed of 14% of Asian patients and 18% of Pineal gland tumors were Black patients. Schwannomas, similarly to meningiomas are driven largely by NF2 gene mutations. Nevertheless, schwannomas showed a lower proportion of Black patients with only 6% of all patients. In contrast, the Asian group represented 13% of all schwannoma cases. These associations should be evaluated in future studies. Notably, a type of tumor missing in our analysis is a germ cell tumor. Germ cell tumors show a very high inflammatory cell content with only scattered tumor cells and are not suitable for molecular analysis using DNA methylation profiling.
Discussion
There is a paucity of CNS tumor data with both racial and molecular annotation. Notably, both CBTRUS and SEER databases show a lack of data for multiple tumor categories and ethnicities. For example, the most recent CBTRUS report does not include any Asian patients in molecular categories of IDH mutant or wild-type glioma, medulloblastoma, or ependymoma. Furthermore, both databases include CNS tumor terminology that is outdated and does not reflect advances in molecular classification of many subtypes of primary CNS tumors and the most recent WHO classification scheme.4 For example, the most recent CBTRUS report includes only Shh, Wnt, and non-Wnt/non-Shh medulloblastoma categories, and only RELA subtype of ependymoma, omitting PFA, PFB, and Yap subtypes. Accurate molecular data are paramount to deciphering the association between race and specific tumor types to guide epidemiological studies and prevention. DNA methylation-based classification has emerged as a robust and reproducible pan-CNS molecular assay, reducing diagnostic errors.9 To the best of our knowledge, our study presents the largest pan-CNS dataset of molecularly annotated tumors with race information and is the first study linking DNA methylation-based classification with race. Using a uniquely diverse CNS tumor cohort from the same geographic area, we show that there is a difference in racial composition both in main molecular groups as well as subgroups of CNS tumors. Current research shows no conclusive evidence that CNS tumor incidence is associated with lifestyle characteristics such as smoking, drug use, alcohol consumption, or dietary choices. Therefore, analyses of racial background provide an important novel insight into the origin of CNS tumors. There have been previous correlations between race and ethnicity and health outcomes and predisposition to disease, including within brain tumors.1,19
Despite extensive diversity, our cohort is still somewhat less diverse than the overall New York City demographic breakdown. According to the United States Census Bureau, 40% of the New York City population is White, 23% is Black, and 14% is Asian,20 in contrast with our cohort, which is 82% White, 10% Black, and 8% Asian. In the future, this can be improved with genotyping analyses which could decrease the number of patients in the Unknown category and inclusion of patients from other hospitals with higher representation of racial minorities. Nevertheless, while some studies have shown limitations in access to molecular diagnostics for patients from racial minorities,21 our institution performs DNA methylation profiling for all patients with primary CNS tumors9 eliminating a selection bias in this study.
In contrast with the previously published work that relied on data from the SEER database, we show that Blacks show the highest prevalence of low-grade gliomas.22 This difference may be due to the different subgroups or lack of molecular benchmarking in other studies compared to our study utilizing advanced molecular testing following the most recent WHO guidelines.
In medulloblastoma, race/ethnicity has been identified as a predictor of brain tumor rates, even when controlled for confounding by socioeconomic status.23 Our data further supports this conclusion, as we found an increase in representation of the White group and a decrease in representation of the Asian group within the medulloblastoma group.
Previous studies have reported a higher incidence of pituitary adenomas in the Black group, which has been found to be independent of socioeconomic status but did not report any difference in incidence between the White and Asian groups.24,25 Our study confirms an increase in incidence in the Black group, and furthermore identifies the overrepresentation of the FSH/LH subgroup. Furthermore, we show an increased incidence in the Asian group, and a decreased incidence in the White group. Our data suggest that these variations may be even more granular and different racial groups may have predisposition to different hormonal subgroups of pituitary adenoma, although this requires additional studies.
IDH mutant gliomas have been studied globally, and show different incidence rates across countries in comparison to IDH wild-type gliomas, including the United States, Europe, and Asia.26 Since these areas have different racial/ethnic breakdowns, the difference in incidence rate may be tied to underlying racial predispositions for this entity.
Similar to previous work, we show an increased incidence of meningiomas among Black patients, but also show an increase in representation of the Black group in one particular subgroup, Benign 3, which has not previously been characterized.27,28
Our data confirm that Whites have a higher prevalence of GBM.29–31 However, we also demonstrate that there may be differences in molecular subgroups with Asian patients showing enrichment for two molecular subgroups of GBM. Other subgroups need to be evaluated in larger studies.
In this study, we utilized self-reported race/ethnicity data to group patients into broader racial groups. While self-reported race/ethnicity data is not as accurate a method as germline genotyping to determine race/ethnicity, it represents the best source of race/ethnicity information for the cohort of CNS tumors without available normal DNA samples. Some studies have shown that DNA methylation profiling can be used for baseline ethnicity determination.32 However, our analysis of tumor DNA methylation showed that it is not sufficiently accurate to distinguish between different races/ethnicities. While it can distinguish White and Black patients, it does not reliably distinguish between White and Asian (data not shown). Therefore, DNA methylation currently cannot be recommended as a substitute for genotyping studies.
Furthermore, there are currently almost 120 molecular subtypes of primary CNS tumors. Therefore, for most subtypes of less common primary CNS tumors, we currently do not have enough cases to draw conclusions about race and molecular subtypes. Our study shows that collaborative efforts across multiple institutions are necessary to elucidate the association between race and many rare molecular subtypes.
Lastly, although we have the benefit of being located in New York City, with a very diverse catchment area, our results may not be representative of every racial group in every geographic area. Further studies should be conducted in other areas around the United States and the world to elucidate globally applicable trends and how race/ethnicity contributes to the development of specific molecular subtypes.
Race and ethnicity emerge from our study as important variables with significant differences in the incidence of particular CNS tumors and molecular subgroups. However, the mechanism of prevalence of different molecular subtypes remains to be elucidated. The extent to which this phenomenon is caused by genetic differences versus social disparities or environment requires further examination. Nonetheless, examination of these differences through race and ethnicity at the genetic level can confer important information as to differences in the incidence of brain tumor types across race and ethnicity to identify additional risk factors and help guide prevention.
Supplementary Material
Contributor Information
Camila S Fang, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Wanyi Wang, Department of Biostatistics, NYU School of Global Public Health, New York, New York, USA.
Chanel Schroff, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Misha Movahed-Ezazi, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Varshini Vasudevaraja, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Jonathan Serrano, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Erik P Sulman, Brain and Spine Tumor Center, Laura and Isaac Perlmutter Cancer Center, NYU Langone Health, New York, New York, USA; Department of Radiation Oncology, NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
John G Golfinos, Brain and Spine Tumor Center, Laura and Isaac Perlmutter Cancer Center, NYU Langone Health, New York, New York, USA; Department of Neurosurgery, NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Daniel Orringer, Department of Neurosurgery, NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Kristyn Galbraith, Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Yang Feng, Department of Biostatistics, NYU School of Global Public Health, New York, New York, USA.
Matija Snuderl, Brain and Spine Tumor Center, Laura and Isaac Perlmutter Cancer Center, NYU Langone Health, New York, New York, USA; Department of Pathology NYU Langone Health and NYU Grossman School of Medicine, New York, New York, USA.
Funding
The study was supported by the Friedberg Charitable Foundation, Gray Family Foundation, Sohn Conference Foundation, Making Headway Foundation, and by NIH grants R01-CA226527, R56-NS122987, and R01-NS122987.
Conflict of interest statement
M.S. is scientific advisor and shareholder of Heidelberg Epignostix and Halo Dx, and a scientific advisor of Arima Genomics, and InnoSIGN, and received research funding from Lilly USA. Other authors declare no conflict of interest.
Authorship statement
Study concept and design: C.S.F. and M.S.; Acquisition of cases and data collection: K.G., M.M.E., M.S., E.P.S., J.G.G., and D.O.; Experiments: C.S. and K.G.; Analysis of data: C.S.F., V.V., J.S., W.W., Y.F., and M.S.; Manuscript Review: C.S.F., M.S., S.W., and Y.F.; Wrote Manuscript: C.S.F. and M.S.; All authors read and approved the final paper.
Data availability
No new data were generated for this research.
References
- 1. Barnholtz-Sloan JS, Ostrom QT, Cote D.. Epidemiology of brain tumors. Neurol Clin. 2018;36(3):395–419. [DOI] [PubMed] [Google Scholar]
- 2. Miller KD, Ostrom QT, Kruchko C, et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin. 2021;71(5):381–406. [DOI] [PubMed] [Google Scholar]
- 3. Ostrom QT, Fahmideh MA, Cote DJ, et al. Risk factors for childhood and adult primary brain tumors. Neuro-Oncology. 2019;21(11):1357–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Organisation mondiale de la santé, Centre international de recherche sur le cancer, editors. Central nervous system tumours. 5th ed. Lyon: International agency for research on cancer; 2021. [Google Scholar]
- 5. Özdemir BC, Dotto G-P.. Racial differences in cancer susceptibility and survival: More than the color of the skin? Trends Cancer 2017;3(3):181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ostrom QT, Price M, Neff C, et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 2023;25(12 suppl 2):iv1–iv99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capper D, Stichel D, Sahm F, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: The Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galbraith K, Vasudevaraja V, Serrano J, et al. Clinical utility of whole-genome DNA methylation profiling as a primary molecular diagnostic assay for central nervous system tumors-A prospective study and guidelines for clinical testing. Neurooncol. Adv.. 2023;5(1):vdad076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Race and National Origin [Internet]. National Institutes of Health (NIH). 2022. https://www.nih.gov/nih-style-guide/race-national-origin. Accessed April 21, 2023. [Google Scholar]
- 12. Serrano J, Snuderl M.. Whole genome dna methylation analysis of human glioblastoma using illumina beadArrays [Internet]. In: Placantonakis DG, editor. Glioblastoma: Methods and Protocols. New York, NY: Springer; 2018. p. 31–51. 10.1007/978-1-4939-7659-1_2. Accessed May 30, 2023. [DOI] [PubMed] [Google Scholar]
- 13. Agresti A. A survey of exact inference for contingency tables. Statist Sci. 1992;7(1):131–153. [Google Scholar]
- 14. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 15. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 16. Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hovestadt V, Jones DTW, Picelli S, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510(7506):537–541. [DOI] [PubMed] [Google Scholar]
- 18. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostrom QT, Price M, Neff C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro-Oncology. 2022;25(5):v1–v95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. U.S. Census Bureau QuickFacts: New York city, New York [Internet]. https://www.census.gov/quickfacts/newyorkcitynewyork. Accessed Apr 25, 2023. [Google Scholar]
- 21. Khoury MJ, Bowen S, Dotson WD, et al. Health equity in the implementation of genomics and precision medicine: A public health imperative. Genet Med. 2022;24(8):1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cao J, Yan W, Zhan Z, Hong X, Yan H.. Epidemiology and risk stratification of low-grade gliomas in the United States, 2004-2019: A competing-risk regression model for survival analysis. Front Oncol. 2023;13:13. https://www.frontiersin.org/articles/10.3389/fonc.2023.1079597. Accessed May 18,2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muskens IS, Feng Q, Francis SS, et al. Pediatric glioma and medulloblastoma risk and population demographics: A Poisson regression analysis. Neurooncol. Adv.. 2020;2(1):vdaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castellanos LE, Gutierrez C, Smith T, Laws ER, Iorgulescu JB.. Epidemiology of common and uncommon adult pituitary tumors in the U.S. according to the 2017 World Health Organization classification. Pituitary. 2022;25(1):201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghaffari – Rafi A, Mehdizadeh R, Ghaffari-Rafi S, et al. Demographic and socioeconomic disparities of pituitary adenomas and carcinomas in the United States. J Clin Neurosci. 2022;98:96–103. [DOI] [PubMed] [Google Scholar]
- 26. Ang SYL, Lee L, See AAQ, et al. Incidence of biomarkers in high-grade gliomas and their impact on survival in a diverse SouthEast Asian cohort - A population-based study. BMC Cancer. 2020;20:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhala S, Stewart DR, Kennerley V, et al. Incidence of benign meningiomas in the united states: current and future trends. JNCI Cancer Spectr. 2021;5(3):pkab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cote DJ, Wang R, Morimoto LM, et al. Birth characteristics and risk of meningioma in a population-based study in California. Neurooncol. Adv.. 2022;4(1):vdac173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS.. Adult glioma incidence and survival by race or ethnicity in the United States From 2000 to 2014. JAMA Oncol. 2018;4(9):1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patel NP, Lyon KA, Huang JH.. The effect of race on the prognosis of the glioblastoma patient: A brief review. Neurol Res. 2019;41(11):967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thakkar JP, Dolecek TA, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23(10):1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan V, Price EM, Del Gobbo G, et al. Accurate ethnicity prediction from placental DNA methylation data. Epigenetics Chromatin 2019;12(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated for this research.