Abstract
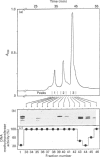
Within the H1 histone family, only some fractions enriched in the H1e-c variants are effective in causing a marked inhibition, in vitro, of enzymic DNA methylation and, in gel retardation and Southwestern blot experiments, in binding double-stranded (ds) CpG-rich oligonucleotides. Both the 6-CpG ds-oligonucleotide and the DNA purified from chromatin fractions enriched in 'CpG islands' are good competitors for the binding of H1e-c to 6-meCpG ds-oligonucleotide. Because of their ability to bind any DNA sequence and to suppress the enzymic methylation in any sequence containing CpG dinucleotides, these particular H1 variants could play some role in maintaining linker DNA at low methylation levels and even in preserving the unmethylated state of the CpG-rich islands which characterize the promoter regions of housekeeping genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Davis T., Fulton J., Kirk D., Qureshi M., Burdon R. H. Eukaryotic DNA methylase--properties and action on native DNA and chromatin. Curr Top Microbiol Immunol. 1984;108:142–156. [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Antequera F., Bird A. CpG islands. EXS. 1993;64:169–185. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- Baubichon-Cortay H., Mallet L., Denoroy L., Roux B. Histone H1a subtype presents structural differences compared to other histone H1 subtypes. Evidence for a specific motif in the C-terminal domain. Biochim Biophys Acta. 1992 Jul 31;1122(2):167–177. doi: 10.1016/0167-4838(92)90320-d. [DOI] [PubMed] [Google Scholar]
- Bavykin S., Srebreva L., Banchev T., Tsanev R., Zlatanova J., Mirzabekov A. Histone H1 deposition and histone-DNA interactions in replicating chromatin. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3918–3922. doi: 10.1073/pnas.90.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., McComas W. W., Weissbach A. DNA methylation: sequences flanking C-G pairs modulate the specificity of the human DNA methylase. Nucleic Acids Res. 1985 May 24;13(10):3479–3494. doi: 10.1093/nar/13.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986 Apr;6(4):1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen B., Steinberg J., Laemmli U. K., Weintraub H. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 1980 Jan 11;8(1):1–20. doi: 10.1093/nar/8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Caiafa P., Attină M., Cacace F., Tomassetti A., Strom R. 5-Methylcytosine levels in nucleosome subpopulations differently involved in gene expression. Biochim Biophys Acta. 1986 Aug 22;867(4):195–200. doi: 10.1016/0167-4781(86)90034-5. [DOI] [PubMed] [Google Scholar]
- Caiafa P., Mastrantonio S., Cacace F., Attinà M., Rispoli M., Strom R. Localization, in human placenta, of the tightly bound form of DNA methylase in the higher order of chromatin organization. Biochim Biophys Acta. 1988 Nov 10;951(1):191–200. doi: 10.1016/0167-4781(88)90040-1. [DOI] [PubMed] [Google Scholar]
- Caiafa P., Reale A., Allegra P., Rispoli M., D'Erme M., Strom R. Histones and DNA methylation in mammalian chromatin. Differential inhibition by histone H1. Biochim Biophys Acta. 1991 Aug 27;1090(1):38–42. doi: 10.1016/0167-4781(91)90034-j. [DOI] [PubMed] [Google Scholar]
- Carotti D., Palitti F., Mastrantonio S., Rispoli M., Strom R., Amato A., Campagnari F., Whitehead E. P. Substrate preferences of human placental DNA methyltransferase investigated with synthetic polydeoxynucleotides. Biochim Biophys Acta. 1986 Mar 26;866(2-3):135–143. doi: 10.1016/0167-4781(86)90110-7. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Cole R. D. Microheterogeneity in H1 histones and its consequences. Int J Pept Protein Res. 1987 Oct;30(4):433–449. doi: 10.1111/j.1399-3011.1987.tb03352.x. [DOI] [PubMed] [Google Scholar]
- D'Erme M., Santoro R., Allegra P., Reale A., Marenzi S., Strom R., Caiafa P. Inhibition of CpG methylation in linker DNA by H1 histone. Biochim Biophys Acta. 1993 May 28;1173(2):209–216. doi: 10.1016/0167-4781(93)90183-e. [DOI] [PubMed] [Google Scholar]
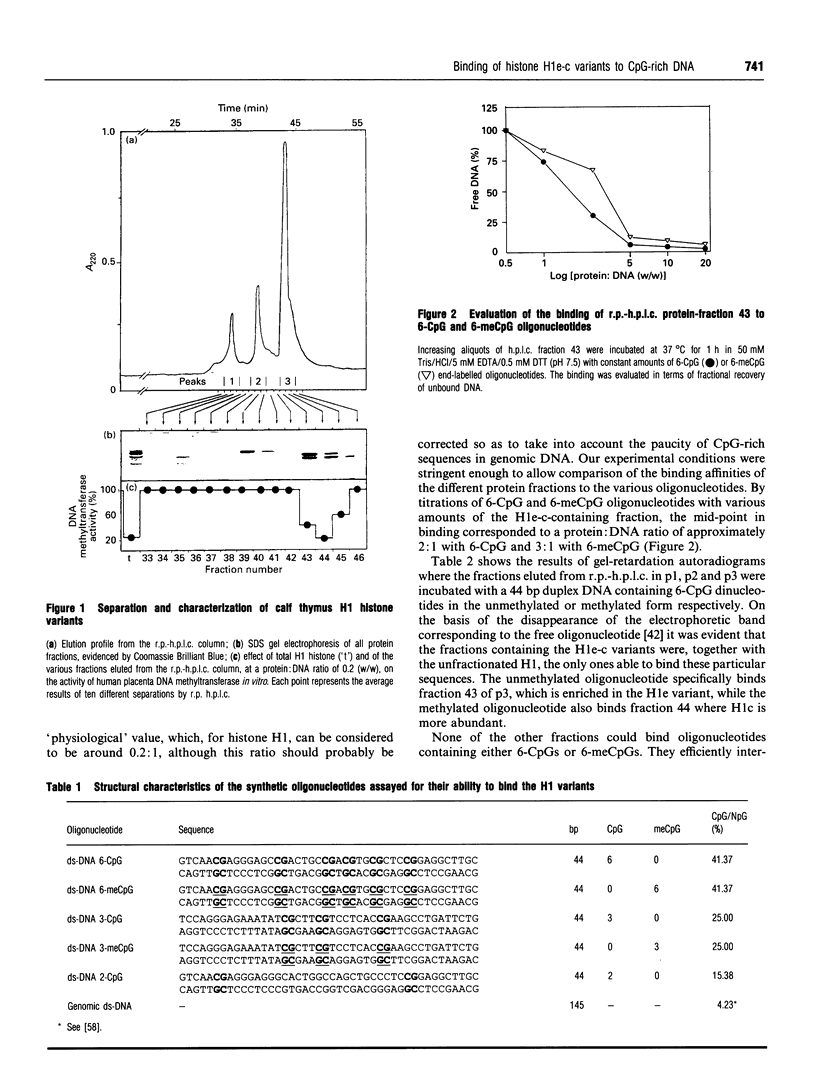
- Davis T., Rinaldi A., Clark L., Adams R. L. Methylation of chromatin in vitro. Biochim Biophys Acta. 1986 May 5;866(4):233–241. doi: 10.1016/0167-4781(86)90048-5. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Faraone-Mennella M. R., D'Erme M., Quesada P., Caiafa P., Farina B. Histone-induced condensation of rat testis chromatin: testis-specific H1t versus somatic H1 variants. Biochem Biophys Res Commun. 1994 Jan 14;198(1):32–39. doi: 10.1006/bbrc.1994.1005. [DOI] [PubMed] [Google Scholar]
- Frank D., Keshet I., Shani M., Levine A., Razin A., Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991 May 16;351(6323):239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Giancotti V., Bandiera A., Ciani L., Santoro D., Crane-Robinson C., Goodwin G. H., Boiocchi M., Dolcetti R., Casetta B. High-mobility-group (HMG) proteins and histone H1 subtypes expression in normal and tumor tissues of mouse. Eur J Biochem. 1993 Apr 15;213(2):825–832. doi: 10.1111/j.1432-1033.1993.tb17825.x. [DOI] [PubMed] [Google Scholar]
- Higurashi M., Cole R. D. The combination of DNA methylation and H1 histone binding inhibits the action of a restriction nuclease on plasmid DNA. J Biol Chem. 1991 May 5;266(13):8619–8625. [PubMed] [Google Scholar]
- Huang H. C., Cole R. D. The distribution of H1 histone is nonuniform in chromatin and correlates with different degrees of condensation. J Biol Chem. 1984 Nov 25;259(22):14237–14242. [PubMed] [Google Scholar]
- Izaurralde E., Käs E., Laemmli U. K. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J Mol Biol. 1989 Dec 5;210(3):573–585. doi: 10.1016/0022-2836(89)90133-2. [DOI] [PubMed] [Google Scholar]
- Jost J. P., Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lennox R. W., Cohen L. H. The histone H1 complements of dividing and nondividing cells of the mouse. J Biol Chem. 1983 Jan 10;258(1):262–268. [PubMed] [Google Scholar]
- Lennox R. W. Differences in evolutionary stability among mammalian H1 subtypes. Implications for the roles of H1 subtypes in chromatin. J Biol Chem. 1984 Jan 10;259(1):669–672. [PubMed] [Google Scholar]
- Levine A., Yeivin A., Ben-Asher E., Aloni Y., Razin A. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J Biol Chem. 1993 Oct 15;268(29):21754–21759. [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Liao L. W., Cole R. D. Condensation of dinucleosomes by individual subfractions of H1 histone. J Biol Chem. 1981 Oct 10;256(19):10124–10128. [PubMed] [Google Scholar]
- Liao L. W., Cole R. D. Differences among H1 histone subfractions in binding to linear and superhelical DNA. Sedimentation velocity studies. J Biol Chem. 1981 Nov 10;256(21):11145–11150. [PubMed] [Google Scholar]
- Liao L. W., Cole R. D. Differences among subfractions of H1 histone in their interactions with linear and superhelical DNA. Circular dichroism. J Biol Chem. 1981 Jul 10;256(13):6751–6755. [PubMed] [Google Scholar]
- Lindner H., Helliger W., Puschendorf B. Separation of rat tissue histone H1 subtypes by reverse-phase h.p.l.c. Identification and assignment to a standard H1 nomenclature. Biochem J. 1990 Jul 15;269(2):359–363. doi: 10.1042/bj2690359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pehrson J. R., Cole R. D. Histone H1 subfractions and H10 turnover at different rates in nondividing cells. Biochemistry. 1982 Feb 2;21(3):456–460. doi: 10.1021/bi00532a006. [DOI] [PubMed] [Google Scholar]
- Quesada P., Farina B., Jones R. Poly(ADP-ribosylation) of nuclear proteins in rat testis correlates with active spermatogenesis. Biochim Biophys Acta. 1989 Mar 1;1007(2):167–175. doi: 10.1016/0167-4781(89)90035-3. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., D'Erme M., Reale A., Strom R., Caiafa P. Effect of H1 histone isoforms on the methylation of single- or double-stranded DNA. Biochem Biophys Res Commun. 1993 Jan 15;190(1):86–91. doi: 10.1006/bbrc.1993.1014. [DOI] [PubMed] [Google Scholar]
- Schulze E., Trieschmann L., Schulze B., Schmidt E. R., Pitzel S., Zechel K., Grossbach U. Structural and functional differences between histone H1 sequence variants with differential intranuclear distribution. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2481–2485. doi: 10.1073/pnas.90.6.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Szyf M., Tanigawa G., McCarthy P. L., Jr A DNA signal from the Thy-1 gene defines de novo methylation patterns in embryonic stem cells. Mol Cell Biol. 1990 Aug;10(8):4396–4400. doi: 10.1128/mcb.10.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C., Bolden A., Nalin C. M., Weissbach A. In vitro methylation of the 5'-flanking regions of the mouse beta-globin gene. J Biol Chem. 1987 Aug 15;262(23):11057–11063. [PubMed] [Google Scholar]
- Wellman S. E., Sittman D. B., Chaires J. B. Preferential binding of H1e histone to GC-rich DNA. Biochemistry. 1994 Jan 11;33(1):384–388. doi: 10.1021/bi00167a049. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990 Jul;15(7):273–276. doi: 10.1016/0968-0004(90)90053-e. [DOI] [PubMed] [Google Scholar]