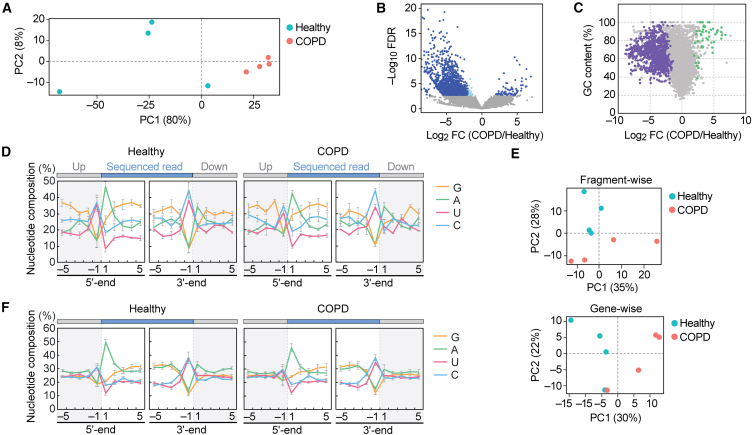
Figure 4.
Analysis of plasma rRFs and mRFs
(A) PCA of rRFs.
(B) Volcano plot for rRFs obtained from DESeq2. The light blue dots indicate a fold change level lower than -2-fold or higher than 2-fold, while the navy-blue dots represent FDR of less than 0.05.
(C) The GC contents of each rRF were plotted against their log2 fold change values. The rRFs with an average count among eight libraries lower than 10 were excluded, and 48 upregulated rRFs and 879 downregulated rRFs with FDR <0.05 are depicted in green and purple, respectively.
(D) Nucleotide composition around the 5′- and 3′-ends of the rRFs. This analysis was performed based on the number of reads, thus reflecting the abundance of each sequence read. A dashed line separates upstream and downstream positions for the 5′- and 3′-ends, representing the cleavage site that generates rRFs (the regions outside of rRF are colored in gray).
(E) PCA of mRFs. PCA was performed using either fragment counts (upper panel) or mRNA ID base (lower panel).
(F) Nucleotide composition around the 5′- and 3′-ends of the mRFs.