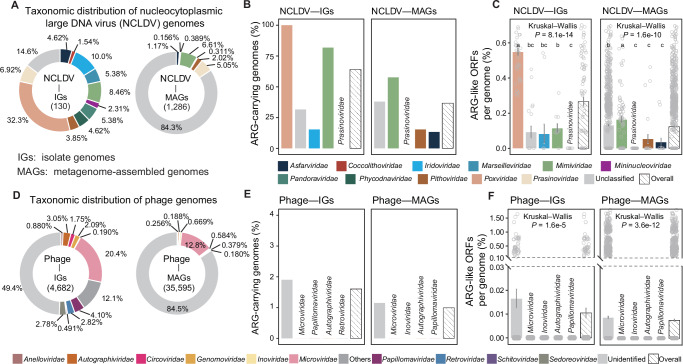
Fig. 1. Taxonomic overview of viral genomes and their antibiotic resistance gene (ARG) carriage.
Taxonomic distribution of the genomes of (A) nucleocytoplasmic large DNA viruses (NCLDVs) and (D) phages analyzed in this study. For NCLDVs, the 11 currently known families are shown. For phages, the 10 most abundant families (according to the number of genomes in individual families) are displayed, and the rest families are referred to as “Others”. Possibility of ARG-carriage in (B) NCLDVs and (E) phages. Genomic potential of ARG-carriage of (C) NCLDVs and (F) phages. For clarity, only the most abundant four families, the unclassified genomes (as a whole), and the overall patterns are displayed in (B–F), with data on all viral families being presented in Supplementary Data 7 and 8. Viral family names are displayed above the bars where colors cannot be clearly recognized. Lower-case letters above the bars in (C) and (F) represent significantly different groups assessed with two-sided Wilcoxon rank-sum test, and P-values indicate the overall difference among all families assessed with Kruskal–Wallis test. Data presented in (C) and (F) were mean values ± standard error of the mean (SEM). Each data point represents an individual genome in the corresponding group. The number (n) of genomes in each group can be found in Supplementary Data 7 for (C) and Supplementary Data 8 for (F). The y-axis was truncated to zoom in on values below 0.03% in (F). ORF is the abbreviation for open reading frame, IG is for isolate genome, and MAG is for metagenome-assembled genome. Source data are provided as a Source Data file on Github107.