Abstract
Small open reading frames (smORFs) shorter than 100 codons are widespread and perform essential roles in microorganisms, where they encode proteins active in several cell functions, including signal pathways, stress response, and antibacterial activities. However, the ecology, distribution and role of small proteins in the global microbiome remain unknown. Here, we construct a global microbial smORFs catalog (GMSC) derived from 63,410 publicly available metagenomes across 75 distinct habitats and 87,920 high-quality isolate genomes. GMSC contains 965 million non-redundant smORFs with comprehensive annotations. We find that archaea harbor more smORFs proportionally than bacteria. We moreover provide a tool called GMSC-mapper to identify and annotate small proteins from microbial (meta)genomes. Overall, this publicly-available resource demonstrates the immense and underexplored diversity of small proteins.
Subject terms: Microbial genetics, Protein databases, Microbial ecology
Here, the authors built a non-redundant catalogue of nearly 1 billion putative small proteins from the global microbiome as a publicly-available resource, and highlight how some highly prevalent and evolutionarily conserved sequences lack functional annotation.
Introduction
Small open reading frames (smORFs) are found in all three domains of life, estimated as 5–10% of annotated genes1–3. Small proteins encoded by smORFs have been reported to perform key functions in microbial cells4–8 and have been found involved in transcription to regulate gene expression9, to stabilize large protein complexes10, in signaling transduction pathways11, regulation of transporters12, sporulation13,14, photosynthesis15, and response to environmental cues16. In addition, small proteins can also perform antibacterial activities17 or compose toxin/antitoxin (TA) systems18,19.
However, small proteins have been neglected in (meta)genomics-based global studies of the microbiome20,21 due to the difficulty in reliably identifying smORFs using genomic information alone22,23. Advances in Ribo-Seq24 and proteogenomics methods25,26 combined with comparative genomics methods27,28 have enabled the discovery of an increasing number of small proteins in various microorganisms29–32. For example, a recent systematic study revealed 4539 novel conserved small protein families of the human microbiome33, 30% of which are predicted to encode transmembrane or secreted proteins. However, most of the studies focusing on smORFs approach isolated microorganisms and specific environments. The functional and ecological understanding of microbial smORFs at a global scale across different habitats is still very limited.
Here, we use the principle that repeated independent observations of the same small protein (or minor variations thereof) minimize the likelihood of false positive smORF predictions and construct a global microbial smORFs catalog (GMSC) derived from 63,410 assembled metagenomes from the SPIRE database21 and 87,920 isolate genomes from the ProGenomes2 database34. In the catalog, we provide comprehensive annotation containing taxonomy classification, habitat assignment, quality assessment, conserved domain (CD) annotation, and predicted cellular localization. In addition, the catalog can be used as a reference to annotate (meta)genomes as the presence of homologs reduces the probability that false positives are reported. To facilitate this, we developed a tool named GMSC-mapper, which additionally provides users with information about the distribution of any matching smORFs across taxonomy, habitats, and geography. Thus, our catalog and associated tools can be used to study the presence, prevalence, distribution, and potential ecological roles of smORFs on a global scale, and provide new insights into how these molecules work within microorganisms.
Results
The global microbial smORFs catalog comprises 965 million smORFs
The global microbial smORFs catalog (GMSC) was derived from 63,410 publicly available assembled metagenomes spanning multiple habitats worldwide from the SPIRE database21 and 87,920 high-quality isolate microbial genomes from the ProGenomes2 database34 (Fig. 1a, Supplementary Data 1). From the assembled contigs, we used the modified version of Prodigal35 in Macrel36 to predict open reading frames (ORFs) with a minimum length of 30 nucleotides (see Methods). The ORFs encoding small proteins (here defined as those up to 100 amino acids) were considered small ORFs (smORFs).
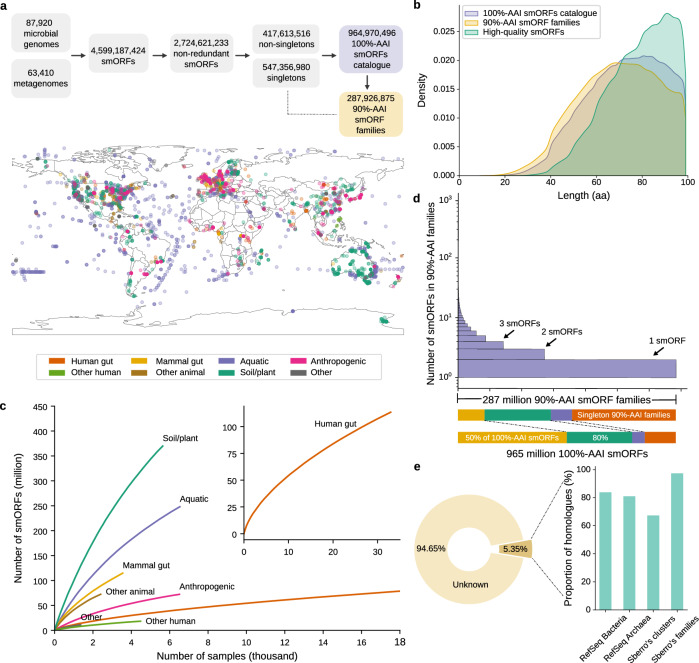
Fig. 1. Global Microbial smORFs Catalog (GMSC).
a ORFs (open reading frames) were predicted from contigs from 63,410 assembled metagenomes from the SPIRE database and 87,920 microbial genomes from the ProGenomes2 database. The ORFs with at most 300 bps were considered smORFs. In total, 4,599,187,424 smORFs were predicted, of which 99.25% originated in metagenomes and 0.75% originated in microbial genomes. The number of smORFs was reduced to 2,724,621,233 by removing redundancy at 100% amino-acid identity (AAI) and 100% coverage. We further clustered the non-redundant smORFs into 287,926,875 clusters at a 90% amino-acid identity (AAI) cutoff (Methods). b Small proteins encoded by smORFs range in length from 9 to 99 amino acids. Sequences that pass all in silico quality tests and contain at least one piece of experimental evidence are considered high-quality predictions (Methods). c Shown are gene accumulation curves per habitat, showing how sampling affects the discovery of smORFs (see also Supplementary Fig. 2a). d The largest 90%-AAI smORF family contains 4577 sequences. The size of 90%-AAI smORF families exhibits a long tail distribution, and 47.5% of families consist of only one sequence, accounting for fewer than 15% of the total GMSC smORFs. A small fraction of large families account for the majority of GMSC smORFs (12.2% of families contain 50% of smORFs). e Only 5.35% of smORFs in the GMSC have a homologous sequence in another sequence catalog (Methods). On the other hand, more than 80% of bacterial and archaeal small proteins from the RefSeq database have a homolog in our catalog. Although only 67.3% of the 444,054 small protein clusters from the Sberro human microbiome dataset are homologous to a protein in our catalog, most of their clusters without homologous sequences only contain one sequence. Among the 4539 conserved small protein families from the Sberro human microbiome dataset, 97.4% of them are homologous to our catalog.
In total, after collapsing smORFs coding for identical amino acid sequences, we obtained 2,724,621,233 smORFs. A large majority (84.7%) were singleton sequences. To reduce the incidence of false positives36, we focused first on the 417 million non-singleton sequences. We hierarchically clustered these non-singleton smORFs at 90% amino-acid identity and 90% coverage, which resulted in 287,926,875 clusters, which we will henceforth refer to as families. Then, we constructed the smORFs catalog, which contains both non-singletons as well as any singleton that matches a family representative at 90% amino-acid identity and 90% coverage (rescued singletons, see Methods). The final smORF catalog contains 964,970,496 smORFs.
The samples in our dataset had been previously manually curated into 75 habitats21, which we further grouped into 8 broad categories: mammal gut, anthropogenic, other-human, other-animal, aquatic, human gut, soil/plant, and other (Methods, Supplementary Data 2). Despite the large number of samples we have collected, rarefaction analysis indicates that smORF diversity is far from covered (Fig. 1c; Supplementary Fig. 2a).
Approximately half of GMSC families consist of only one sequence, but the size distribution of families is long-tailed, so the largest 12.2% of families already cover half of the 100AA smORFs (Fig. 1d).
43 million smORFs are high-quality
Predicting smORFs can result in a high rate of false positives. Thus, in addition to discarding non-homolog singleton predictions, we performed several in silico quality tests including estimating coding potential of families using RNAcode37 and additionally matching genomic predictions to publicly available metatranscriptomic and metaproteomics data (see Methods). In total, 43,642,695 (4.5%) of the smORFs pass all in silico quality tests and have at least one match in transcriptional or translational data. We henceforth refer to these as high-quality predictions (Supplementary Figs. 3 and 4).
To assess the comprehensiveness of our catalog, we matched small proteins encoded by GMSC smORFs to the RefSeq database38 and previously published human microbiome small protein family datasets33. Only 5.3% of smORFs in our catalog are homologous to these previously reported small proteins (Fig. 1e). On the other hand, our catalog contains more than 80% of these reference datasets. For smORFs of high-quality predictions, a higher proportion (8.7%) show homology with these reference datasets, but they only cover circa 20% of the reference datasets (Supplementary Fig. 5a). Hence the high-quality predictions produce a large number of novel small proteins with high confidence that are not present in other reference datasets, but as the available transcriptome and metaproteome datasets are limited, discarding non-high-quality predictions would result in a large loss of coverage.
To explore the functions undertaken by the small proteins encoded by the smORFs in our catalog, we searched the small protein families against the Conserved Domain Database (CDD)39 using RPS-BLAST40,41. Only 6.1% of small protein families containing 86,694,259 smORFs (8.98%) were assigned CDD domains, compared to 35.2% of canonical-length proteins (greater than 100 amino acids)20. As expected, smORFs in high-quality predictions are twice as likely to be assigned a CDD domain (18.8%, P value < , hypergeometric test).
Even conserved small proteins lack functional annotations
Using MMSeqs2 taxonomy42 we predicted the taxonomic origin of contigs and transferred that prediction to the smORFs (Methods). This process returned a prediction for 81.6% of the 100AA smORFs, with more than half (56.9%) being assigned to a genus or species (Fig. 2a). Note that we used the GTDB database43, which does not include phage or microeukaryotes.
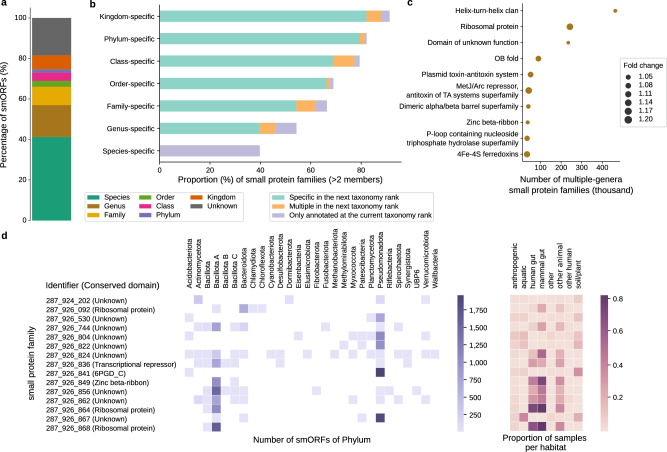
Fig. 2. Taxonomic and functional annotation of small proteins.
a Predicting taxonomy for the contigs and genomes from which smORFs originate (Methods) resulted in a taxonomic assignment for 81.6% of smORFs (56.9% of smORFs at genus or species level). b When only families with >2 members were considered (96,721,815 families), there are three cases at each taxonomic rank. For example, considering the rank of class, a small protein family is annotated to a particular taxonomic class if all its members are annotated as belonging to that class (unannotated smORFs being ignored). We further distinguish three cases, namely whether its members are (i, marked specific in the next taxonomic rank) all be annotated to the same order (as order is the next taxonomic rank), (ii, marked multiple in the next taxonomic rank) annotated to different orders within that class, or (iii, marked only annotated at the current taxonomic rank) not annotated to any order. Other ranks are treated analogously (until we reach the level of species). c The enrichment of Pfam domains in small protein families present in multiple genera compared to the entire families with over two members (P value < 0.05, Hypergeometric Test, corrected by Bonferroni). Pfam domains were grouped by Pfam domain clans. Fold change is the ratio of the Pfam proportion of small protein families which present in multiple genera to the Pfam proportion of the entire families with over two members. d The Pfam annotation of small protein families that exist in multiple phyla, spanning >100 species and distributed across all the eight broad habitat categories (mammal gut, anthropogenic, other-human, other-animal, aquatic, human gut, soil/plant, and other).
We next investigated the taxonomic breadth and conservation of smORFs28,44. Of the 96,721,815 small protein families with at least three members, more than half of them (52,550,829) are genus-specific (Fig. 2b). Among these genus-specific families, most are species-specific, accounting for 39.7% of the families included in the analysis.
Although in some cases, smORFs may be present in plasmids and other mobile elements, we reasoned that multi-genus families would be especially likely to be present in multiple habitats and involved in critical cellular functions33. As expected, multi-genus families are more common in multiple habitats than the entire set of families with at least three members even when differences in family size distributions are taken into account, but the difference is not large (61.8% vs. 57.5%; P value < , due to the large number of datapoints, hypergeometric test). Furthermore, we traced the conserved Pfam domains of small protein families45 (Supplementary Data 4). Multi-genus families are annotated with Pfam domains at a higher rate than the background of all families with at least three members (9.91% vs 8.15%; P value < , hypergeometric test). Nonetheless, it is noteworthy that the vast majority have no detected Pfam domain and that a further 9.5% of those annotated, were annotated with Pfam domains of unknown functions (Fig. 2c).
We then focused on conserved families present in multiple phyla. We found a total of 2437 multi-phylum families present across all 8 broad habitat categories (Supplementary Data 5). Of these, only 752 families were annotated with Pfam CDs, of which 268 (35.6%) were associated with ribosomal proteins and 99 (13.2%) belonged to the Helix-turn-helix clan (Fig. 2d).
Archaea harbor more smORFs proportionally than bacteria
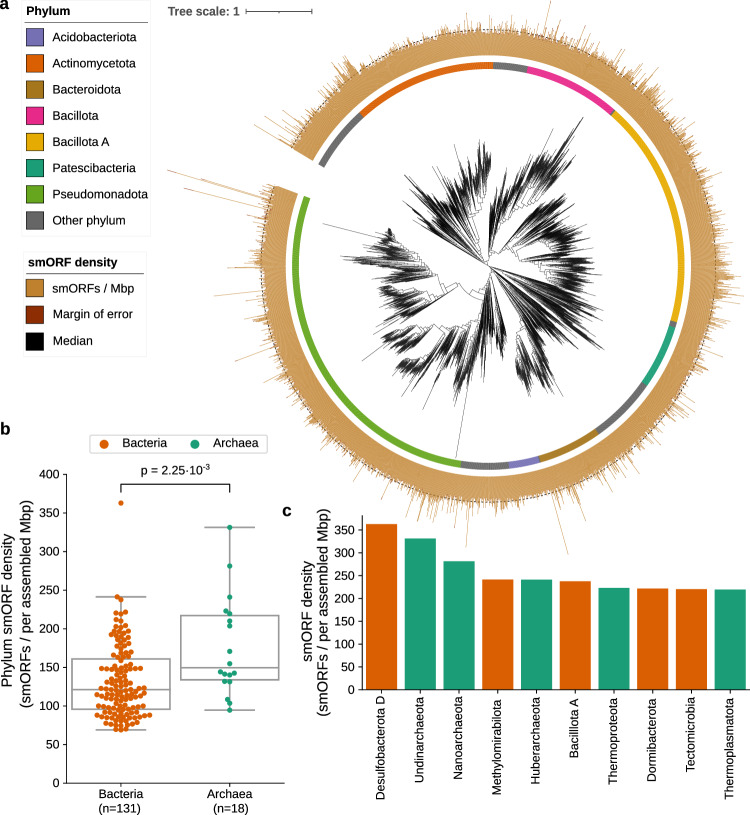
To investigate the presence of smORFs in different microorganisms without sampling bias, we calculated the number of redundant smORFs per megabase pairs (Mbp) of assembled contigs, also named the density of smORFs32,46.
Most of the genera with the highest density come from Pseudomonadota, Bacillota A, Actinomycetota, Bacillota, and Bacteroidota (Fig. 3a). However, when considering the density of phyla as a whole, interestingly, we found the density of archaeal phyla is higher than bacterial ones ( = ; Fig. 3b). Of the ten phyla with the highest smORF density, half are archaeal, despite the fact that only 18 archaeal phyla contained enough data to be analyzed compared to 131 bacterial ones (Fig. 3c, Supplementary Data 6). The phyla that produce the most smORFs per Mbp are Desulfobacterota D (362.87 smORFs per Mbp), Undinarchaeota (331.35 smORFs per Mbp), Nanoarchaeota (281.34 smORFs per Mbp), Methylomirabilota (241.37 smORFs per Mbp), and Huberarchaeota (241.05 smORFs per Mbp).
Fig. 3. Archaea harbor more smORFs than bacteria.
a Shown is the smORFs density distribution for the top 3000 bacterial genera with the highest density (brown bars, confidence interval of 95% shown as dark brown bars). Most of the densest genera are from Pseudomonadota, Bacillota A, and Actinomycetota. For reference, the black dashed line represents the median smORFs density for the presented genera. b Calculating the smORFs density of each phylum, the density of archaea is significantly higher than that of bacteria. Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers) as well as outliers (single points) that lie within 1.5 IQRs of the lower and upper quartile. P values shown are from the Mann–Whitney test (two-sided). c The top 10 phyla with the highest smORF density are shown.
Differences in functions for archaeal and bacterial small proteins
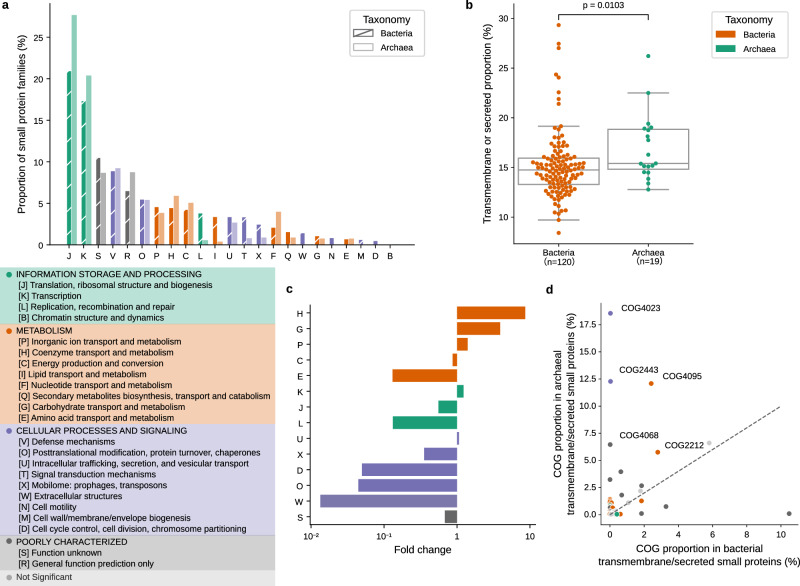
Given the higher densities of smORFs in Archaea, we investigated the functions and properties in archaeal and bacterial small proteins encoded by smORFs47. We compared the archaeal and bacterial small protein families with COG48 annotation. Only 1.72% of the families are annotated with COGs, of which 4,747,223 families are from bacteria and 202,825 families are from archaea. The COG classes that belong to Information storage and processing account for the largest proportion of small proteins in both bacteria and archaea (Fig. 4a), which is consistent with that found by Wang et al.44. However, circa 17% of small proteins in bacteria and archaea are still annotated as COG classes which are poorly characterized.
Fig. 4. Differences in functional prediction for archaeal and bacterial small proteins.
a The COG distribution of archaeal and bacterial small proteins is shown. b Archaea contain a higher fraction of transmembrane or secreted small proteins than bacteria (calculated per phylum). Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers) as well as outliers (single points) that lie within 1.5 IQRs of the lower and upper quartile. P values shown are from the Mann–Whitney Test (two-sided). c Shown is the difference in the proportion of COG class in archaeal transmembrane or secreted small proteins versus bacterial transmembrane or secreted small proteins. The fold change is the ratio of proportions. The P values were calculated using Fisher’s exact test (two-sided) and adjusted by Bonferroni correction. d Dots represent 43 COGs, which are enriched in archaeal transmembrane or secreted small proteins compared to the archaeal small proteins that are not transmembrane or secreted, as well as bacterial transmembrane or secreted small proteins. The proportion comparison of these 43 COGs between archaeal transmembrane or secreted small proteins and bacterial transmembrane or secreted small proteins is shown.
Small proteins with transmembrane or secreted characteristics may be involved in cell communication7. We explored the transmembrane and secreted small proteins in archaea and bacteria (Methods). 15.3% of the families are predicted to be transmembrane (using TMHMM-2.049) or secreted (using SignalP-5.050), with archaeal families being predicted at a higher rate than bacterial ones to be transmembrane or secreted ( ≤ 0.0103, Fig. 4b)51.
Furthermore, compared with bacterial transmembrane or secreted small proteins, we found that archaeal transmembrane or secreted small proteins are enriched in COG classes related to the transport and metabolism of coenzymes, carbohydrates, and inorganic ions, besides the intracellular trafficking, secretion, and vesicular transport. In contrast, they are depleted in COG classes related to cellular processes and signaling (P value < 0.05, Fisher’s exact test, multiple tests corrected by Bonferroni, Fig. 4c).
Some COGs were primarily (or even exclusively) present in archaea (as defined by a P value < 0.05, Fisher’s exact test, multiple tests corrected by Bonferroni, Fig. 4d). For example, the COG with the highest proportion in archaea, COG4023 is a preprotein translocase subunit Sec61beta, which is a component of the Sec61/SecYEG protein secretion system. It is found in eukaryotes and archaea and is possibly homologous to the bacterial SecG52.
Identification of smORFs by GMSC-mapper
As mentioned above, smORF predictions are prone to false positives and one strategy for increasing confidence is to find sequences present in multiple genomes (or metagenomes). In this context, our catalog can be a resource whereby users with a single sample (or a small number of samples) use it as a reference to obtain high-quality predicted smORFs. For this usage, we provide a tool called GMSC-mapper (Fig. 5a).
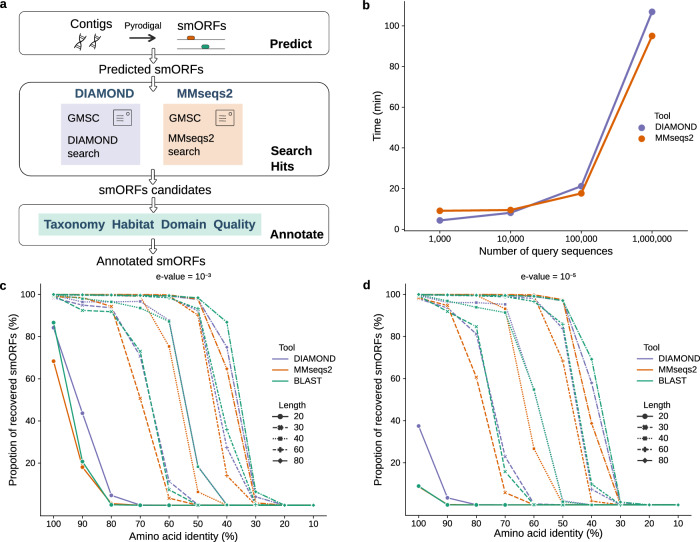
Fig. 5. Workflow and benchmark of GMSC-mapper.
a GMSC-mapper uses Pyrodigal to predict small proteins with <100 amino acids from contigs. Users can alternatively provide smORF or protein sequences directly, skipping the initial step of gene prediction. DIAMOND or MMseqs2 are used for finding homologs within GMSC. In the end, GMSC-mapper combines all alignment hits and provides detailed annotations of small proteins. b Time cost tests were performed among different numbers of input sequences from 1000 to 1,000,000 using DIAMOND and MMseqs2 (Methods). We compared the number of recovered sequences with different lengths (20, 30, 40, 60, and 80 amino acids) at different amino acid identities from 10% to 100% using DIAMOND, MMseqs2, and BLAST (Methods). The recovered number is influenced by the E value cutoff used ( in c and in d).
GMSC-mapper performs de novo prediction and annotation of small proteins encoded by smORFs in user-provided genomes or assembled metagenomes (Methods). For this, it first uses Pyrodigal35,53 to predict small proteins from assembled contigs and then it uses DIAMOND54 or MMseqs255 to align these predictions against the GMSC. To minimize computational resource usage, the GMSC-mapper only searches family representatives, but it returns the set of matching smORFs and the annotation of the matches (e.g., habitat and taxonomy) as well as links to GMSC identifiers.
We compared DIAMOND to MMseqs2 for this task and observed that DIAMOND is faster than MMseqs2 when the number of query sequences is below 10,000, while MMseqs2 is slightly faster than DIAMOND when the number of queries is above 10,000 (Fig. 5b). In addition, we compared the number of recovered sequences (Fig. 5c) with either of these tools or BLAST56, by randomly modifying sequences in the catalog and aligning these modified versions back to the catalog of family representatives. All three tools can find a high-identity match if it is present in the database. With increasing sequence size, these tools can match more distant homologous sequences. In this case, DIAMOND achieves almost the same sensitivity as BLAST and is superior to MMseqs257.
However, independently of the method used, when the sequences are too short (20 amino acids), the rate of recovery decreases drastically. Fundamentally, for short sequences in a large database, even an identical match has a high likelihood of arising by chance58,59. This will manifest itself in a high E value60 for true positives, making it impossible to distinguish false and true positive matches based on sequence comparisons alone. Therefore, while the use of a higher E value threshold will recover a larger fraction of true matches (>80% recovered with DIAMOND using compared to circa 40% using , see Fig. 5c, d), the false discovery rate (FDR) will also increase58.
Discussion
Here, we constructed the global microbial smORFs catalog (GMSCv1, in its first version), which contains ~1 billion smORF sequences, of which 43 million are high-quality predictions, representing a large increase in the number of smORF sequences previously reported and serving as a resource for the microbiome research community. For each smORF and small protein family, we provide comprehensive annotations, including taxonomy, habitats, and CDs. Previously, most of the widely studied microbial small proteins were accidentally discovered in isolated and cultured bacterial species5. The large-scale discovery of small proteins has made great progress in recent years. Sberro et al.33 conducted the characterization of conserved small proteins in the human microbiome, revealing their potential various functions. In our work, we have expanded the discovery of small proteins to 75 distinct habitats worldwide. In our catalog, only a small fraction are homologous to reference small protein datasets, with the vast majority of the novel small proteins being found in non-human-associated habitats (Supplementary Fig. 5b). On the other hand, it encompasses most of the known small proteins in either the RefSeq database or in families discovered recently (NMPfamsDB61 and FesNov families28). When comparing with small protein databases that focus on eukaryotic organisms, such as smProt262, OpenProt2.063, and sORF.org64, the overlap is minimal (Supplementary Fig. 5c).
A major difficulty in finding biologically functional smORFs is that false positive predictions are common. One of the underlying principles in our efforts is that finding identical or highly similar sequences in multiple samples increases the likelihood of a true prediction. Therefore, we discarded singleton predictions in our data. This principle also underlies the GMSC-mapper tool, which enables users to find matches from their datasets in GMSCv1.
As previously done33, we have only conducted RNAcode37 on small protein families with at least eight members to identify smORFs families with transcription signatures. This approach may, however, fail to identify some rapidly evolving functional smORFs. In addition, given the limited size and number of existing datasets of metatranscriptomes, (meta)Ribo-Seq and metaproteomes, the high-quality predictions are expected to underestimate the true diversity.
Computing approaches and concepts developed over decades for longer proteins do not necessarily work well for small sequences. For example, for the alignment of very short sequences, the minimum achievable E value will be lower bounded60. Even an identical match will obtain a relatively high E value as short identical matches can occur by chance. Furthermore, traditional databases lack small proteins, so functional assignment by orthology or with HMMs only returns a prediction for a minute fraction of all small proteins. We lack functional predictions for most small proteins in our dataset, even for those small protein families that are ubiquitous. Similarly, tools for predicting whether proteins are transmembrane or secreted are not optimized for small proteins and our results should be interpreted in this context. In particular, when we compared results between bacteria and archaea, we implicitly assumed that the methods have similar error rates in these two domains, but this may not be the case. In related work, we used machine learning36 to identify candidate antimicrobial peptides (AMPs) from the GMSC46. However, functional prediction for small proteins remains an open challenge, open to new approaches.
Overall, our resource shows the immense and underexplored diversity of small proteins across different habitats and taxonomy, and highlights the gaps in our scientific knowledge, while constituting a resource for the research community.
Methods
Collection of global metagenomic datasets and prediction of smORFs
In total, 63,410 publicly available global assembled metagenomes from the SPIRE database21 collection were used. Briefly, the assembled metagenomes have been generated through the following methods: publicly available data (as of 1 January 2020) were downloaded from the European Nucleotide Archive (ENA) and short reads that were at least 60 bps after trimming positions with quality <2565 were assembled into contigs using MEGAHIT 1.2.966. Additionally, we downloaded 87,920 high-quality isolate microbial genomes from the ProGenomes2 database34.
We then used the modified version of Prodigal35 in Macrel 0.536 to predict ORFs ≥ 30 base pairs (bps) on the assembled contigs as well as those from Progenomes2 database. This version of Prodigal uses the same algorithm as the standard version of Prodigal, but with a lower limit on the size of genes. We used command line parameters to only predict closed genes, to not predict genes with N as a base, to perform a full motif scan, in metagenomics mode (-c -m -n -p meta). The ORFs encoding small proteins (here defined as those up to 100 amino acids) were considered smORFs.
We recorded the habitats of smORFs according to their source samples using the habitat microontology introduced in SPIRE database21. We further grouped the habitats into 8 broad categories: mammal gut, anthropogenic, other-human, other-animal, aquatic, human gut, soil/plant, and other. We used GeoPandas67 to present geographic coordinates of samples.
Non-redundant smORFs catalog construction and method validation
All the smORFs were first deduplicated at 100% amino-acid identity and 100% coverage. Then we hierarchically clustered the non-singletons at 90% amino-acid identity and 90% coverage using Linclust55,68 with the following parameters: -c 0.9, –min-seq-id 0.9. Linclust is a single-linkage approach, whereby sequences are clustered together if they share a common representative with candidate representatives being chosen heuristically.
Of these clusters, 47.5% contain a single sequence (singleton clusters). To rule out the possibility that this was due to the fact that Linclust55,68 is a heuristic method that is not specifically designed for short sequences, we estimated the rate of false negatives (i.e., sequences that were marked as singleton even though they should have been clustered with another one). We aligned a randomly selected 1000 singleton clusters against the representative sequences of non-singleton clusters (i.e., those containing ≥2 sequences) using SWIPE69 with the following parameters: -a 18 -m ‘8 std qcovs’ -p 1. The alignment threshold was E value < , identity ≥90%, and coverage ≥90% (Supplementary Fig. 1a).
In addition, to estimate the rate of false positive clusterings (sequences that were assigned to a cluster even though they do not share the required identity with the cluster representative), 1000 sequences were randomly selected and aligned against the representative sequences of their clusters using SWIPE69 with the following parameters: -a 18 -m ‘8 std qcovs’ -p 1. The alignment threshold was E value < , identity ≥90%, and coverage ≥90% (Supplementary Fig. 1b).
When clustering, we initially discarded the singletons because singletons are enriched in artifactual smORFs36. However, we considered that singletons that are homologous to larger clusters should not be discarded as the homology itself provides further evidence of biological relevance. Therefore, we aligned singletons to the representative sequences of clusters with 90% sequence identity and 90% coverage using DIAMOND54 using parameters: -e –id 90 -b 12 -c 1 –query-cover 90 –subject-cover 90. We combined the homolog singletons and the non-singleton sequences identified earlier and termed them the smORFs catalog containing 964,970,496 smORFs.
Sample-based smORFs rarefaction curves
Samples were randomly permuted 24 times to calculate the total number of non-redundant smORFs captured as the number of samples increased. We took the average across the permutations as the final estimate.
Quality control of the catalog
We conducted several in silico quality tests and matched genomic predictions to other publicly available experimental data.
A smORF predicted at the start of a contig that is not preceded by an in-frame STOP codon risks being a false positive originating from an interrupted fragment. Therefore, we checked for the presence of an upstream in-frame STOP. For smORFs without an upstream in-frame STOP, however, we could not determine whether there were other genes present upstream of them (Supplementary Fig. 3a).
To avoid spurious smORFs, we used HMMSearch70 with the --cut_ga option to search smORFs against the AntiFam 7.0 database71, which contains a series of confirmed spurious protein families.
We used RNAcode37, a tool to predict the coding potential of sequences based on evolutionary signatures, to identify the coding potential of 25,744,932 smORF families containing ≥8 sequences. The smORF families with P value < 0.05 were considered to have coding potential, as in a previous study33 (Supplementary Fig. 4a).
Furthermore, we searched for evidence that these smORFs are transcribed and/or translated. For this step, we downloaded 221 publicly available metatranscriptomic datasets from the NCBI database paired with the metagenomic samples we used in our catalog (Supplementary Data 3). These samples are from the human gut, peat, plant, and symbionts. To keep the procedure computationally feasible, we mapped reads against the representative sequences of smORF families by BWA72. Then we used NGLess65 with ‘unique_only’ for the ‘multiple’ argument of the count built-in function to only count uniquely mapped inserts. A smORF family was considered to have transcriptional evidence if its representative has reads mapped to it in at least 2 samples (Supplementary Fig. 4b). Furthermore, we mapped reads against the smORFs in paired metagenomic and metatranscriptome samples, separately. On average, 58.6% of the smORFs in each paired sample are mapped.
We downloaded 142 publicly available Ribo-Seq datasets from the NCBI database (Supplementary Data 3). We also mapped reads against representative sequences of smORF families by BWA72. Then we used NGLess65 with ‘unique_only’ for the ‘multiple’ argument of the count built-in function to only count uniquely mapped inserts. A smORF family was considered to have translation evidence only if its representative has reads mapped to it in at least 2 samples (Supplementary Fig. 4c).
Moreover, we downloaded peptide datasets from 108 metaproteomic projects from the PRIDE database73 (Supplementary Data 3). We matched GMSC smORFs to the identified peptides of each project. If the total k-mer coverage of peptides on a smORF is greater than 50%, then the smORF is considered translated and detected, as in a previous study74 (Supplementary Fig. 4d).
Sequences that passed all in silico tests above as well as matching transcriptional or translational data were regarded as high-quality predictions.
Comparison with reference small protein datasets
We downloaded bacterial and archaeal protein sequences from RefSeq in March 202338, consensus sequences of NMPFamsDB61 and sequences for each FESNov gene family28. The sequences with fewer than 100 amino acids are considered small proteins, and redundancy was subsequently removed with 100% amino-acid identity and 100% coverage. A total of 16,333,323 bacterial small proteins, 368,769 archaeal small proteins from RefSeq, 56,786 small proteins from NMPFamsDB, and 630,375 small proteins from FESNov families were included in the comparison. We also included the 444,053 small protein clusters and 4539 conserved small protein families from Sberro’s human microbiome study33. We compared our smORFs catalog to these datasets using DIAMOND with the ‘–more-sensitive’ mode, retaining significant hits (E value < ). In addition, we compared our smORFs catalog with small protein sequences provided in current small protein database mainly about eukaryotic organisms. We downloaded small proteins from human, mouse, yeast, rat, E. coli, C. elegans, fruitfly, zebrafish, and small proteins from LiteratureMining, KnownDatabase, and MSfragments from SmProt2 database62; small proteins from human, mouse, rat, zebrafish, fruitfly, C. elegans of sORF.org database64; and all predicted refprots, altprots, and isoforms sequences with all annotations from human, chimp, rat, mouse, zebrafish, fruitfly, C. elegans, and yeast from OpenProt2.0 database63. After filtering small proteins by length and removing redundancy as above, 788,586 small proteins from SmProt2 database, 4,377,422 small proteins from sORF.org database, and 1,781,907 small proteins from OpenProt2.0 database were included in the comparison. As above, we compared our smORFs catalog to these datasets using DIAMOND with the ‘–more-sensitive’ mode, retaining significant hits (E value < ).
Conserved domain annotation
We downloaded the CDD39 from ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/little_endian/Cdd_LE.tar.gz in September 2022, which contains models from CD curated at NCBI, Pfam45, SMART75, COGs48, PRK76, and TIGRFAMs77. All the representative sequences of small protein families were searched against the CDD by RPS-BLAST40,41. In order to establish a comparison baseline, we additionally randomly selected 10,000 prokaryotic proteins from the global microbial gene catalog v1.020 and searched them against the CDD by RPS-BLAST40,41. Hits with an E-value maximum of 0.01 and at least 80% of coverage of PSSM’s length were retained and considered significant. Pfam accessions were grouped by Pfam clan78 or the first phrase before the comma in their short description.
Taxonomic annotation and taxonomic breadth analysis
The taxonomy of assembled contigs encoding the small proteins was annotated using MMseqs2 taxonomy42 against the GTDB database43 release r95. However, in figures and text, we used updated taxon names (e.g., Bacillota instead of Firmicutes). We characterized the taxonomy of predicted smORFs based on the taxonomy of contigs and microbial genomes34 from which the smORFs were predicted. We subsequently assigned taxonomy for GMSC smORFs and families using the lowest common ancestor, ignoring the un-assigned ranks to make them more specific.
The small protein families with at least three members were subsequently used to perform taxonomic breadth analysis. Each family was classified according to (i) whether it is single or multi-habitat; (ii) whether it is single or multi-genus; and (iii) whether it is annotated with a Pfam domain45. Multi-genus families are more common in multiple habitats than the entire families (61.8% vs. 52.0%; P value < , hypergeometric test). Multi-genus families are annotated with Pfam domains at a higher rate (9.91% vs 7.52%; P value < , hypergeometric test). As these results could have been confounded by differences in family size distributions, we randomly downsampled the data to keep the same number of families at each size between multi-genus families and the whole families. In that case (as presented in the main text), the difference was 61.8% vs. 57.5% (P value < , hypergeometric test) for the proportion of families in multiple habitats and 9.91% vs. 8.15% (P value < , hypergeometric test) for the proportion of Pfam annotated families.
Density calculation
The density of smORFs was defined as = / , where is the number of redundant smORFs and L is the assembled megabase pairs (Mbps)32,46. The density was calculated by summing all assembled base pairs for contigs assigned to each taxonomic rank. We assume a scenario where the starting positions of smORFs in an assembled large contig are independent and uniformly random. Therefore, the standard sample proportion error was calculated as and was used to calculate the margin of error at a 95% confidence interval (Z = 1.96). We did not further consider the calculated values with a margin of error >10%.
Cellular localization prediction
To detect potential transmembrane proteins, we ran TMHMM-2.049 on the representative sequences of small protein families. Then, to identify potentially secreted small proteins, we used SignalP-5.050 on the representative sequences of small protein families. For families classified as archaea, we used ‘-org arch’, while for the others we combined the outputs of ‘-org gram + ’ and ‘-org gram-’ modes.
Construction and evaluation of GMSC-mapper
GMSC-mapper supports assembled contigs, smORF sequences, or protein sequences as inputs. It uses Pyrodigal35,53, which is a faster implementation of the Prodigal algorithm, to predict ORFs potentially coding for small proteins (those with fewer than 100 amino acids) from contigs. Gene prediction is skipped when inputs are smORF or protein sequences. Then DIAMOND54 or MMseqs255 are used for homologous alignment against GMSC. Finally, it combines all the alignment hits information and provides detailed annotation of small proteins.
To determine the optimal default sensitivity mode, we tested different sensitivity parameters for DIAMOND and MMseqs2. We aligned 10,000 randomly selected sequences back to the family representatives and counted the number of recovered sequences while monitoring the computational time. We use the “–sensitive” mode as the default sensitivity parameter for DIAMOND, which provides the best balance between sensitivity and speed. The use of more-sensitive modes resulted in little or almost no increase in the number of recovered sequences, but a substantial increase in time usage. For MMseqs2, we keep the original default sensitivity parameter (5.7) considering that the number of recovered sequences and the time both increase with the increase of sensitivity (Supplementary Fig. 6a-d).
We then tested the time costs among different numbers of input sequences using the “–sensitive” mode of DIAMOND and the default sensitivity parameter (5.7) of MMseqs2. GMSC-mapper can annotate 100,000 input sequences in approximately 20 minutes with 20 threads.
Furthermore, we compared the number of recovered sequences with different identities using different alignment tools. We randomly selected and mutated 10,000 sequences of different lengths (20, 30, 40, 60, and 80) from the family representatives, with different identities from 10% to 90%. We aligned them back to the family representatives using DIAMOND, MMseqs2, and BLAST56, respectively. When the query sequence and the target sequence are the same, we consider them as the recovered sequences.
Timing measurements were performed using a server equipped with an AMD EPYC 7763 64-Core processor and 2TB of RAM memory.
Statistics and reproducibility
Statistical analyses were carried out in Python 3.8.5, using Pandas79 1.1.3, NumPy80 1.24.4, and SciPy81 1.10.1. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
GMSC web resource
GMSC webserver is hosted at the address https://gmsc.big-data-biology.org, where an implementation of GMSC-mapper can be accessed. The website implementation is based on Elm-Lang. The API implementation is based on Python.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the National Natural Science Foundation of China (T2225015, 61932008) (L.P.C., Z.X.M.), Shanghai Science and Technology Commission Program (23JS1410100) (L.P.C., Z.X.M.), National Key R&D Program of China (2023YFF1204800, 2020YFA0712403) (L.P.C., Z.X.M.), Key Science and Technology Project of Hainan Province (ZDYF2024SHFZ058) (Z.X.M.), Major Project of Guangzhou National Laboratory (GZNL2024A01003) (Z.X.M.), Lingang Laboratory & National Key Laboratory of Human Factors Engineering Joint Grant (LG-TKN-202203-01) (Z.X.M.), and the Australian Research Council (grant FT230100724). We thank Ben Woodcroft (Queensland University of Technology) for helpful comments on a previous version of the manuscript.
Author contributions
L.P.C. conceptualized and designed the study. Y.D., C.D.S.J., T.S.B.S., A.F., L.P.C., M.K. curated the data. Y.D., C.D.S.J., B.L.S.D., and C.Z. analyzed and visualized the data. L.P.C., X.M.Z., and P.B. supervised the project. Y.D. and L.P.C. wrote the original draft. All authors reviewed and contributed to the manuscript.
Peer review
Peer review information
Nature Communications thanks Zachary Ardern, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Global metagenomic data are publicly available at the ENA. The accession numbers for samples and studies are listed in Supplementary Data 1. Microbial genomes are publicly available in the Progenomes2 database. The global microbial smORFs catalog (GMSC) resource has been deposited in Zenodo under 10.5281/zenodo.7944370. The resource is freely available at https://gmsc.big-data-biology.org. Users can query small protein sequences by using GMSC-mapper through the web interface or select their interesting small proteins by habitats and taxonomy.
Code availability
The codes used to generate and analyze the global microbial smORFs catalog (GMSC) are available at https://github.com/BigDataBiology/Duan2024__GMSCv1_Construction_And_Analysis, archived at Zenodo under 10.5281/zenodo.13119583. GMSC-mapper is open source and at https://github.com/BigDataBiology/GMSC-mapper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xing-Ming Zhao, Email: xmzhao@fudan.edu.cn.
Luis Pedro Coelho, Email: luispedro@big-data-biology.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51894-6.
References
- 1.Kastenmayer, J. P. et al. Functional genomics of genes with small open reading frames (sORFs) in S. Cerevisiae. Genome Res.16, 365–373 (2006). 10.1101/gr.4355406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su, M., Ling, Y., Yu, J., Wu, J. & Xiao, J. Small proteins: untapped area of potential biological importance. Front. Genet.4, 286 (2013). 10.3389/fgene.2013.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pueyo, J. I., Magny, E. G. & Couso, J. P. New peptides under the s(ORF)ace of the genome. Trends Biochem. Sci.41, 665–678 (2016). 10.1016/j.tibs.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 4.Hobbs, E. C., Fontaine, F., Yin, X. & Storz, G. An expanding universe of small proteins. Curr. Opin. Microbiol.14, 167–173 (2011). 10.1016/j.mib.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storz, G., Wolf, Y. I. & Ramamurthi, K. S. Small proteins can no longer be ignored. Annu. Rev. Biochem.83, 753–777 (2014). 10.1146/annurev-biochem-070611-102400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval, M. & Cossart, P. Small bacterial and phagic proteins: an updated view on a rapidly moving field. Curr. Opin. Microbiol.39, 81–88 (2017). 10.1016/j.mib.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 7.Yadavalli, S. S. & Yuan, J. Bacterial small membrane proteins: the swiss army knife of regulators at the lipid bilayer. J. Bacteriol.204, e00344–21 (2022). 10.1128/JB.00344-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weidenbach, K., Gutt, M., Cassidy, L., Chibani, C. & Schmitz, R. A. Small proteins in archaea, a mainly unexplored world. J. Bacteriol.204, e00313–e00321 (2022). 10.1128/JB.00313-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altieri, A. S. et al. A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp. Nucleic Acids Res.44, 6232–6241 (2016). 10.1093/nar/gkw351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaßel, M., Möllenkamp, T., Puppe, W. & Altendorf, K. The KdpF subunit is part of the K+-translocating Kdp complex of escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem.274, 37901–37907 (1999). 10.1074/jbc.274.53.37901 [DOI] [PubMed] [Google Scholar]
- 11.Salazar, M. E., Podgornaia, A. I. & Laub, M. T. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol. Microbiol.102, 430–445 (2016). 10.1111/mmi.13471 [DOI] [PubMed] [Google Scholar]
- 12.Lloyd, C. R., Park, S., Fei, J. & Vanderpool, C. K. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J. Bacteriol.199, e00869–16 (2017). 10.1128/JB.00869-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutting, S. et al. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J. Bacteriol.179, 5534–5542 (1997). 10.1128/jb.179.17.5534-5542.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmalisch, M. et al. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol.192, 5402–5412 (2010). 10.1128/JB.00534-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanOrsdel, C. E. et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol.195, 3640–3650 (2013). 10.1128/JB.00324-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alix, E. & Blanc-Potard, A.-B. Hydrophobic peptides: novel regulators within bacterial membrane: regulatory membrane peptides in bacteria. Mol. Microbiol.72, 5–11 (2009). 10.1111/j.1365-2958.2009.06626.x [DOI] [PubMed] [Google Scholar]
- 17.Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature540, 280–283 (2016). 10.1038/nature20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilmaerts, D. et al. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. mBio9, e00744–18 (2018). 10.1128/mBio.00744-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unoson, C. & Wagner, E. G. H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli: mode of action of TisB. Mol. Microbiol.70, 258–270 (2008). 10.1111/j.1365-2958.2008.06416.x [DOI] [PubMed] [Google Scholar]
- 20.Coelho, L. P. et al. Towards the biogeography of prokaryotic genes. Nature601, 252–256 (2022). 10.1038/s41586-021-04233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt, T. S. B. et al. SPIRE: a searchable, planetary-scale microbiome REsource. Nucleic Acids Res.52, D777–D783 (2023). [DOI] [PMC free article] [PubMed]
- 22.Gray, T., Storz, G. & Papenfort, K. Small proteins; big questions. J. Bacteriol.204, e00341–21 (2022). 10.1128/JB.00341-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr, M. W., Mao, Y., Storz, G. & Qian, S.-B. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res.48, 1029–1042 (2020). 10.1093/nar/gkz734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aspden, J. L. et al. Extensive translation of small open reading frames revealed by poly-ribo-seq. eLife3, e03528 (2014). 10.7554/eLife.03528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petruschke, H., Anders, J., Stadler, P. F. & Jehmlich, N. & von Bergen, M. Enrichment and identification of small proteins in a simplified human gut microbiome. J. Proteom.213, 103604 (2020). 10.1016/j.jprot.2019.103604 [DOI] [PubMed] [Google Scholar]
- 26.Leong, A. Z.-X. et al. Short open reading frames (sORFs) and microproteins: an update on their identification and validation measures. J. Biomed. Sci.29, 19 (2022). 10.1186/s12929-022-00802-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemm, M. R., Paul, B. J., Schneider, T. D., Storz, G. & Rudd, K. E. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol.70, 1487–1501 (2008). 10.1111/j.1365-2958.2008.06495.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez Del Río, Á. et al. Functional and evolutionary significance of unknown genes from uncultivated taxa. Nature626, 377–384 (2024). 10.1038/s41586-023-06955-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez-Laslop, N., Sharma, C. M., Mankin, A. & Buskirk, A. R. Identifying small open reading frames in prokaryotes with ribosome profiling. J. Bacteriol.204, e00294–21 (2022). 10.1128/JB.00294-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petruschke, H. et al. Discovery of novel community-relevant small proteins in a simplified human intestinal microbiome. Microbiome9, 55 (2021). 10.1186/s40168-020-00981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackowiak, S. D. et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol.16, 179 (2015). 10.1186/s13059-015-0742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fremin, B. J. et al. Thousands of small, novel genes predicted in global phage genomes. Cell Rep.39, 110984 (2022). 10.1016/j.celrep.2022.110984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sberro, H. et al. Large-scale analyses of human microbiomes reveal thousands of small, novel genes. Cell178, 1245–1259.e14 (2019). 10.1016/j.cell.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mende, D. R. et al. proGenomes2: an improved database for accurate and consistent habitat, taxonomic and functional annotations of prokaryotic genomes. Nucleic Acids Res.48, D621–D625 (2019). [DOI] [PMC free article] [PubMed]
- 35.Hyatt, D. et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics11, 119 (2010). 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Júnior, C. D., Pan, S., Zhao, X.-M. & Coelho, L. P. Macrel: Antimicrobial peptide screening in genomes and metagenomes. PeerJ8, e10555 (2020). 10.7717/peerj.10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Washietl, S. et al. RNAcode: Robust discrimination of coding and noncoding regions in comparative sequence data. RNA17, 578–594 (2011). 10.1261/rna.2536111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res.44, D733–D745 (2016). 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J. et al. The conserved domain database in 2023. Nucleic Acids Res.51, D384–D388 (2023). 10.1093/nar/gkac1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer, A. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res.30, 281–283 (2002). 10.1093/nar/30.1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul, S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.25, 3389–3402 (1997). 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirdita, M., Steinegger, M., Breitwieser, F., Söding, J. & Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics37, 3029–3031 (2021). 10.1093/bioinformatics/btab184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res.50, D785–D794 (2022). 10.1093/nar/gkab776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, F. et al. A systematic survey of mini-proteins in bacteria and archaea. PLoS One3, e4027 (2008). 10.1371/journal.pone.0004027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res.49, D412–D419 (2021). 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos-Júnior, C. D. et al. Discovery of antimicrobial peptides in the global microbiome with machine learning. Cell187, 3761–3778.e16 (2024). [DOI] [PubMed]
- 47.Liu, J. & Rost, B. Comparing function and structure between entire proteomes. Protein Sci.10, 1970–1979 (2001). 10.1110/ps.10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galperin, M. Y. et al. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res.49, D274–D281 (2021). 10.1093/nar/gkaa1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol.305, 567–580 (2001). 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 50.Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol.37, 420–423 (2019). 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- 51.Koonin, E. V., Mushegian, A. R., Galperin, M. Y. & Walker, D. R. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol.25, 619–637 (1997). 10.1046/j.1365-2958.1997.4821861.x [DOI] [PubMed] [Google Scholar]
- 52.Kinch, L. N., Saier, M. H. Jr & Grishin, N. V. Sec61β-a component of the archaeal protein secretory system. Trends Biochem. Sci.27, 170–171 (2002). 10.1016/S0968-0004(01)02055-2 [DOI] [PubMed] [Google Scholar]
- 53.Larralde, M. Pyrodigal: Python bindings and interface to Prodigal,an efficient method for gene prediction in prokaryotes. J. Open Source Softw.7, 4296 (2022). 10.21105/joss.04296 [DOI] [Google Scholar]
- 54.Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods18, 366–368 (2021). 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol.35, 1026–1028 (2017). 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- 56.Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics10, 421 (2009). 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernández-Salmerón, J. E. & Moreno-Hagelsieb, G. Progress in quickly finding orthologs as reciprocal best hits: comparing blast, last, diamond and MMseqs2. BMC Genomics21, 741 (2020). 10.1186/s12864-020-07132-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ladoukakis, E., Pereira, V., Magny, E. G., Eyre-Walker, A. & Couso, J. Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol.12, R118 (2011). 10.1186/gb-2011-12-11-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couso, J.-P. & Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol.18, 575–589 (2017). 10.1038/nrm.2017.58 [DOI] [PubMed] [Google Scholar]
- 60.Karlin, S. & Altschul, S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci.87, 2264–2268 (1990). 10.1073/pnas.87.6.2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baltoumas, F. A. et al. NMPFamsDB: a database of novel protein families from microbial metagenomes and metatranscriptomes. Nucleic Acids Res.52, D502–D512 (2024). 10.1093/nar/gkad800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, Y. et al. SmProt: a reliable repository with comprehensive annotation of small proteins identified from ribosome profiling. Genomics Proteom. Bioinformatics19, 602–610 (2021). 10.1016/j.gpb.2021.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leblanc, S. et al. OpenProt 2.0 builds a path to the functional characterization of alternative proteins. Nucleic Acids Res.52, D522–D528 (2024). 10.1093/nar/gkad1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olexiouk, V., Van Criekinge, W. & Menschaert, G. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res.46, D497–D502 (2018). 10.1093/nar/gkx1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coelho, L. P. et al. NG-meta-profiler: fast processing of metagenomes using NGLess, a domain-specific language. Microbiome7, 84 (2019). 10.1186/s40168-019-0684-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics31, 1674–1676 (2015). 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 67.Jordahl, K. et al. Geopandas/geopandas: v0.8.1. 10.5281/zenodo.3946761 (2020).
- 68.Steinegger, M. & Söding, J. Clustering huge protein sequence sets in linear time. Nat. Commun.9, 2542 (2018). 10.1038/s41467-018-04964-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rognes, T. Faster Smith-Waterman database searches with inter-sequence SIMD parallelisation. BMC Bioinformatics12, 221 (2011). 10.1186/1471-2105-12-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Potter, S. C. et al. HMMER web server: 2018 update. Nucleic Acids Res.46, W200–W204 (2018). 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eberhardt, R. Y. et al. AntiFam: a tool to help identify spurious ORFs in protein annotation. Database2012, bas003 (2012). 10.1093/database/bas003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows transform. Bioinformatics25, 1754–1760 (2009). 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma, Y. et al. Identification of antimicrobial peptides from the human gut microbiome using deep learning. Nat. Biotechnol.40, 921–931 (2022). 10.1038/s41587-022-01226-0 [DOI] [PubMed] [Google Scholar]
- 75.Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res.46, D493–D496 (2018). 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klimke, W. et al. The National Center for Biotechnology Information’s protein clusters database. Nucleic Acids Res.37, D216–D223 (2009). 10.1093/nar/gkn734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haft, D. H. et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res.41, D387–D395 (2012). 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finn, R. D. Pfam: clans, web tools and services. Nucleic Acids Res.34, D247–D251 (2006). 10.1093/nar/gkj149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKinney, W. Data structures for statistical computing in Python. In: SCIPY 2010, org.s3-website-us-east-1.amazonaws.com, 56–61 (2010).
- 80.Harris, C. R. et al. Array programming with NumPy. Nature585, 357–362 (2020). 10.1038/s41586-020-2649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Global metagenomic data are publicly available at the ENA. The accession numbers for samples and studies are listed in Supplementary Data 1. Microbial genomes are publicly available in the Progenomes2 database. The global microbial smORFs catalog (GMSC) resource has been deposited in Zenodo under 10.5281/zenodo.7944370. The resource is freely available at https://gmsc.big-data-biology.org. Users can query small protein sequences by using GMSC-mapper through the web interface or select their interesting small proteins by habitats and taxonomy.
The codes used to generate and analyze the global microbial smORFs catalog (GMSC) are available at https://github.com/BigDataBiology/Duan2024__GMSCv1_Construction_And_Analysis, archived at Zenodo under 10.5281/zenodo.13119583. GMSC-mapper is open source and at https://github.com/BigDataBiology/GMSC-mapper.