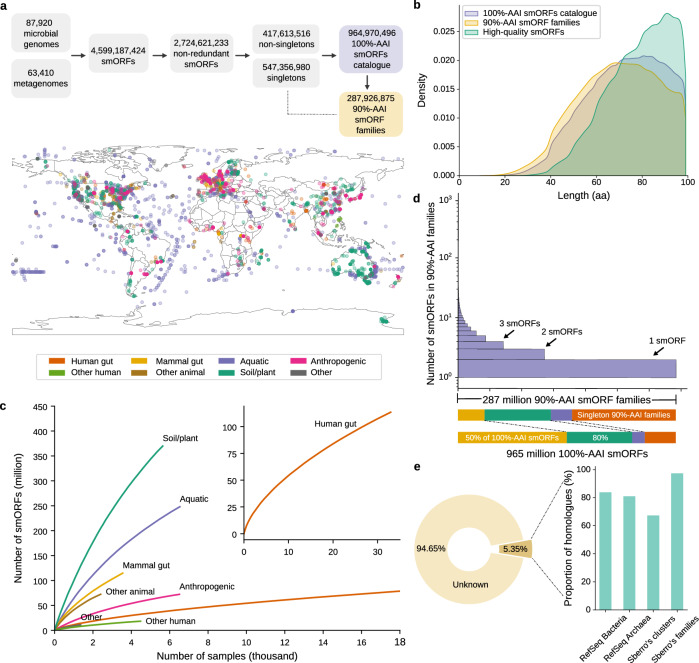
Fig. 1. Global Microbial smORFs Catalog (GMSC).
a ORFs (open reading frames) were predicted from contigs from 63,410 assembled metagenomes from the SPIRE database and 87,920 microbial genomes from the ProGenomes2 database. The ORFs with at most 300 bps were considered smORFs. In total, 4,599,187,424 smORFs were predicted, of which 99.25% originated in metagenomes and 0.75% originated in microbial genomes. The number of smORFs was reduced to 2,724,621,233 by removing redundancy at 100% amino-acid identity (AAI) and 100% coverage. We further clustered the non-redundant smORFs into 287,926,875 clusters at a 90% amino-acid identity (AAI) cutoff (Methods). b Small proteins encoded by smORFs range in length from 9 to 99 amino acids. Sequences that pass all in silico quality tests and contain at least one piece of experimental evidence are considered high-quality predictions (Methods). c Shown are gene accumulation curves per habitat, showing how sampling affects the discovery of smORFs (see also Supplementary Fig. 2a). d The largest 90%-AAI smORF family contains 4577 sequences. The size of 90%-AAI smORF families exhibits a long tail distribution, and 47.5% of families consist of only one sequence, accounting for fewer than 15% of the total GMSC smORFs. A small fraction of large families account for the majority of GMSC smORFs (12.2% of families contain 50% of smORFs). e Only 5.35% of smORFs in the GMSC have a homologous sequence in another sequence catalog (Methods). On the other hand, more than 80% of bacterial and archaeal small proteins from the RefSeq database have a homolog in our catalog. Although only 67.3% of the 444,054 small protein clusters from the Sberro human microbiome dataset are homologous to a protein in our catalog, most of their clusters without homologous sequences only contain one sequence. Among the 4539 conserved small protein families from the Sberro human microbiome dataset, 97.4% of them are homologous to our catalog.