Abstract
Camel milk is a nutrient-rich diet and fermentation affects its nutritional value and probiotic function. In this study, sour camel milk and oat jujube sour camel milk were prepared using fermentation bacteria agent TR1, and the metabolites of camel milk, sour camel milk and oat jujube sour camel milk were detected using a non-targeted metabolomics approach using liquid chromatography-mass spectrometry (LC-MS).The results showed that the partial least squares discriminant analysis (PLS-DA) with 100 % accuracy and good predictive power detected 343 components in positive ion mode and 220 components in negative ion mode. The differential metabolites were mainly organic acids, amino acids, esters, vitamins and other substances contained in camel milk.It showed that there were significant differences in the metabolites of camel milk, sour camel milk and oat jujube sour camel milk. Based on the pathway enrichment analysis of the three dairy products in the KEGG database, 12 metabolic pathways mainly involved in the positive ion mode and 20 metabolic pathways mainly involved in the negative ion mode were identified. The main biochemical metabolic pathways and signal transduction pathways of the differential metabolites of the three dairy products were obtained. This study provides theoretical support for improving the nutritional quality and probiotic function of camel milk and fermented camel milk products and provides a basis for the development of relevant processing technologies and products for camel milk and fermented camel milk.
Keywords: Camel milk, Fermentation bacteria agent TR1, Sour camel milk, Oat jujube sour camel milk, Probiotic function, Non-targeted metabolomics
1. Introduction
Camel milk is a nutritious, medicinal and antibacterial diet popular mainly in Sudan, Arab countries, India, Central Asia and arid regions of western China, such as Inner Mongolia, Gansu and Xinjiang [[1], [2], [3]]. Camel milk is easily absorbed, viscous, hunger-resistant, ethnically distinctive, and meets the nutritional needs of the human body [4,5]. Camel milk has various therapeutic effects due to its composition and is beneficial in treating chronic hepatitis and improving liver function [[6], [7], [8]]. It also contains a high percentage of total salt, calcium, protective proteins and some trace elements such as iron, copper and zinc [[9], [10], [11]]. It has gained widespread attention especially for its low cholesterol content and high vitamin C content [12,13]. Camel milk is the best food for people living in arid and semi-arid regions because of its unique composition and nutritional value [14,15]. Oats are a coarse grain of the grass family and are rich in protein and unsaturated fatty acids such as linoleic and linolenic acids [[16], [17], [18]]. Also, oats are rich in soluble dietary fiber such as vitamins VB1, VB2, VE, VB3, VH, and β-glucan, and rich in minerals such as calcium, phosphorus, iron, zinc, magnesium, and selenium making them highly nutritious [19,20]. Studies have shown that β-glucan in oats can improve immunity and have some anticancer effects [21]. In addition, as a prebiotic, it can be fermented by Bifidobacteria and Lactobacilli in the intestinal tract, promoting the proliferation of these bacteria and thus affecting the intestinal tract [[22], [23], [24]]. The intestinal flora is regulated. Currently, there are many reports on the use of oats to make functional foods, such as oat flour yogurt, wheat yogurt crisps, yak yogurt crisps, and purple potato oat goat milk [25]. Jujube is rich in polysaccharides, ferritin, fat, amino acids, organic acids, various trace elements and vitamins, making it useful for blood, strengthening the spleen and stomach, tonifying the muscles and bones, and relieving fatigue. Relevant studies have shown that it also has anti-aging and anti-cancer effects [26,27]. A series of products have been developed using red dates, among which the development of single and compound red date yogurts continues to heat up, while compound red date yogurts such as red date fruit and vegetable yogurt and red date dietary fiber yogurt [28,29]. It can not only improve the aroma and taste of fermented milk, but also enrich the nutritional composition of various raw materials, which is a promising direction and research hotspot.
The four strains of lactic acid bacteria isolated and identified from natural sour camel milk by our group were found to have high in vitro antioxidant capacity, which were above 86 % for DPPH (1,1-Diphenyl-2-picrylhydrazyl free radical), above 14 % for superoxide radicals, above 98 % for hydroxyl radicals and above 58 % for ABTS (2,2′-Azinobis-3-ethylbenzthiazoline 6-sulphonate).The active substance for scavenging free radicals was extracellular secretion; in vitro cholesterol scavenging rate were above 58 %; extracellular polysaccharide content was below 0.0358 mg/mL [30]. Four strains of lactic acid bacteria and one commercially available bacterium were compounded to make the fermenting agent TR1. Sour camel milk and oat jujube sour camel milk were prepared by fermenting camel milk with TR1, and their nutritional composition, live bacterial count and acetaldehyde were analyzed. It was found that the nutritional composition of the 2 types of sour camel milk obtained by their fermentation changed, with an increase in the number of live bacteria and an increase in the acetaldehyde content. In addition, many active factors with health functions, such as bacteriocins, polysaccharides and vitamins, have been produced, giving camel milk a higher value and making it increasingly important in biomedicine, food industry production and scientific research.
Current research work on sour camel milk has focused on the analysis of microbial community structure, antibacterial, antioxidant, lipid-lowering, and hypoglycemic active substances in traditional fermented camel milk, isolation and active substance characterization of lactic acid bacteria, evaluation of bile salt solubility of lactic acid bacteria, and development of flavored sour camel milk [31].However, the beneficial vital components of camel milk, sour camel milk, and oat jujube sour camel milk and their differences have not been determined, and many unknown nutrients and active ingredients have not been reported.
Metabolomics is mainly the analysis of small molecule metabolites (molecular weight <1000 Da) of substrates and products produced by different metabolic pathways, which not only allows a comprehensive analysis of the changes in metabolite content, but also elucidates the patterns of changes between different metabolites and each treatment condition, thus evaluating the biological responses and effects of different substances [32,33]. Metabolomics has been applied in the study of plant, animal and microbial metabolomes, active substances and chemical components.LC-MS untargeted analysis is a valuable tool in the field of metabolomics, with high speed, sensitivity and relative simplicity of sample preparation compared to other analytical platforms [34].Arapitsas Panagiotis et al. [35]used liquid chromatography-mass spectrometry (LC-MS) to analyze wine metabolomics fingerprints and obtain different chromatograms in positive and negative ion mode. Ji Lei et al. [36] (identified potential biomarkers of human atherosclerotic abdominal aortic aneurysms by untargeted metabolomics and transcriptomics. Luo et al. [37]used ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry to find that fermentation using Scrophularia coronata could enhance the effective active substances of black tea tea broth, and the results of the study could provide some basis for liquid fermentation research.These findings amply confirm the success of untargeted metabolomics in detecting biometabolomics, but metabolic analyses of camel milk as well as beneficial components in fermented camel milk have been rarely reported.
In this study, we used liquid mass spectrometry (LC-MS) non-targeted metabolomics technique to detect all metabolites in camel milk, fermentative bacterium TR1 fermented sour camel milk and oat jujube sour camel milk, and the compositional differences between camel milk, sour camel milk, and oat jujube sour camel milk were analyzed. A combination of multivariate statistical analysis and one-way statistical analysis was used to analyze the types and amounts of metabolites in camel milk, sour camel milk, and oat jujube sour camel milk, and to elucidate the metabolic differences among camel milk, sour camel milk, and oat jujube sour camel milk. The metabolites with significant differences were screened. A preliminary comparative study of camel milk, sour camel milk and oat jujube sour camel milk was carried out to explain the efficacy of camel milk, sour camel milk and oat jujube sour camel milk at a deeper level, and the detected substances can provide some data and theoretical support for the evaluation of the nutritional value of camel milk, sour camel milk, oat jujube sour camel milk and the corresponding probiotic function research.
2. Materials and methods
2.1. Strains, materials and reagents
Fresh camel milk: fresh camel milk samples were collected from 7-year-old Bactrian camels from herders' homes in the Alxa region. Camel milk samples were collected in June 2021, filtered, pasteurized, and stored at −20 °C in the laboratory.
Strains: four lactic acid bacteria strains (Lactobacillus plantarum strain KC28, Lactobacillus plantarum strain K25, Lactobacillus D1501 and Mesentericus SHU1396), preserved in the Laboratory of Biochemistry and Molecular Biology, Inner Mongolia Agricultural University. The strain used in this experiment, Lactobacillus acidophilus 2888 was isolated from the fermentation agent of Beijing Chuanxiu Technology Co. Ltd.
Methanol and acetonitrile (purity not less than 99 %) were from Thermo; methyl tert-butyl ether (purity not less than 99 %) was from Wokai; formic acid, from TCI; ammonium formate (purity not less than 99 %) was from Sigma; all chemical reagents were chromatographically pure grade.
2.2. Instruments and equipment
Liquid Chromatograph and Mass Spectrometer are produced by Thermo Company.
2.3. Methods
2.3.1. Preparation of sour camel milk and oat jujube sour camel milk
Sour camel milk: fermented camel milk prepared by fermenting bacterium TR1 made from Lactobacillus plantarum strain KC28: Lactobacillus plantarum strain K25: Lactobacillus D1501: Mesentericus SHU1396: Lactobacillus acidophilus 2888 in the compound of 2:1:3:5:3. The ratio of camel milk (volume, mL): complex bacterium TR1 (wet weight, g) was 30:1. The fermentation conditions of sour camel milk were 38 °C, left to seal and ferment for 24 h and then shelved in the refrigerator for 24 h after post-ripening in the freezer.
Oat jujube sour camel milk: camel milk is mixed with 1 % glucose, 5 % jujube powder and 3 % oatmeal, pasteurized and fermented under the same fermentation conditions as sour camel milk to prepare oat jujube sour camel milk.
2.3.2. Analysis of physical and chemical indicators
Live bacteria count assay: The live bacteria count was determined by dilution plate coating method, and the number of colonies was recorded with three replicates in each group.
pH measurement: pH was measured before and after fermentation using a pH meter.
Determination of acetaldehyde content: using iodine titration method, acetaldehyde reacts with sodium bisulfite in acidic environment, the reaction product is sodium acetaldehyde bisulfite, oxidation reaction occurs with iodine and the remaining sodium bisulfite, the product of the reaction in the first step under alkaline conditions, sodium acetaldehyde bisulfite will react quantitatively with iodine, able to calculate the content of acetaldehyde in the measured sample according to the titration consumption of iodine.
2.3.3. Determination of nutrient content
Total sugars were determined by the 3,5-dinitrosalicylic acid method, which produces reducing monosaccharides from the hydrolysis of total sugars. The reducing monosaccharides produce brownish-red 3-amino-5-nitrosalicylic acid when heated with 3,5-dinitrosalicylic acid, a compound with a maximum absorption value at 540 nm. The reducing sugars from the hydrolysed sample minus the unhydrolysed sample were used to produce the polysaccharide [38]. The protein was determined by the Coomassie brilliant blue G-250 method. The maximum absorbance value of the brownish red Coomassie brilliant blue G-250 is at 465 nm and its reaction with the protein to form the blue Coomassie brilliant blue G-250-protein complex has a maximum absorbance value at 595 nm. The protein content can be calculated by measuring the increase in the OD595 value [39]. The fat content of the samples was determined using the Gerber method [40]. The lactose content of the samples was determined using the Reducing Sugar method Reducing Sugar This method is based on the fact that lactose readily oxidizes and reduces another compound, and involves the reaction of lactose with a reagent to form a colored compound colorimetrically Measurement of absorbance using a spectrophotometer, and determination of the lactose concentration according to Beer's law, in which the concentration of the colored compound formed is directly proportional to the concentration of lactose present in the sample Acidity was determined by acid-base Acidity is determined by acid-base titration [41].The acidity was determined by the acid-base titration method [42].
2.3.4. LC-MS sample preparation
The camel milk was fermented according to the selected optimal fermenting conditions, from which 1 mL of sample was taken for the next analysis. Before use, organic solvent was pre-cooled in 40 °C. 150 μL of each sample was taken into 2 mL centrifuge tubes; 200 μL of methanol (−20 °C) and 200 μL of methyl tert-butyl ether (MTBE) were added to each tube, shaken for 60 s and centrifuged at 12 000 rpm for 10 min at 4 °C, and the supernatant was filtered through a 0.22 μm membrane to obtain the sample to be tested.
Quality Control (QC) samples were used to balance the next steps of the Chromatography-Mass Spectrometry System and determine the instrument status. Therefore, both experimental samples and QC samples were detected throughout the entire experimental procedure. There were three groups of experimental samples (fresh camel milk group, fermented camel milk group and oat jujube sour camel milk group). Simultaneously, QC samples were prepared, which were a mixture of equal amounts of all experimental samples. For QC samples, 3 technical replicates were performed.
2.3.5. LC-MS spectral acquisition
Chromatographic conditions: The gradient elution was performed on an ACQUITY UPLC® HSS T3 1.8 μm (2.1 × 150 mm) column with an autosampler set at 8 °C. The mobile phase was positive ion 0.1 % formic acid in water (C) - 0.1 % formic acid in acetonitrile (D); negative ion 5 mM ammonium formate in water (A) - acetonitrile (B). The gradient elution program was 0∼1 min, 2%B/D; 1–9 min, 2 %∼50 % B/D; 9–12 min, 50 %∼98 % B/D; 12∼13.5 min, 98 % B/D; 13.5–14 min, 98 %∼2 % B/D; 14–20 min, 2 % D - positive mode (14–17 min, 2 % B - negative mode) [43].
Mass spectrometry conditions: The instrument uses an electrospray ionization source (ESI) in positive and negative ionization mode with a positive ion spray voltage of 3.50 kV and a negative ion spray voltage of 2.50 kV, a sheath gas flow rate of 50 arb and an auxiliary gas flow rate of 13 arb [[44], [45], [46]]. Sheath gas and auxiliary gas were set at 30 and 10 arbitrary units, respectively [46]. The capillary temperature was 325 °C, a full scan was performed at a resolution of 70 000 with a scan range of 81 to 1000 [47], and secondary lysis was performed using HCD with a collision voltage of 30 eV [48], while dynamic exclusion was used to remove unnecessary MS/MS information.
2.3.6. Data preprocessing
The raw data were converted to mzXML format (xcms input file format) by Proteowizard software; peaksidentification, peak filtration, and peak alignment were performed using R's XCMS package; a data matrix including mass to charge ratio (m/z) and retention time and intensity was obtained. The data matrix including mass to charge ratio (m/z), retention time and peak area (intensity) was obtained; 32009 precursor molecules were obtained in positive ion mode and 11693 in negative ion mode. The data were exported to excel for subsequent analysis [45].
2.3.7. Component structure identification
Metabolites and differential metabolites were identified using the Compound Discovery program by means of exact mass number matching (<10 ppm) and secondary spectrum matching, followed by identification of metabolites and differential metabolites based on MS/MS fragmentation patterns with reference to HMDB, METLIN, Massbank and LipidMaps as well as self-built standards databases, annotation of metabolites to obtain named fractions, and classification.
2.3.8. Screening classification and functional annotation of differential metabolites
The results of the statistical analysis of the experiment were analyzed using SPSS 20.0 software for data analysis, in which the Duncan method was used for one-way ANOVA and the Pearson method was used for correlation analysis. Data were presented as the mean ± SEM and differences of P < 0.05 were considered significant.
Multivariate statistical analysis was used to finally obtain the shared metabolites and differential metabolites. The data were subjected to multivariate statistical analysis (including principal component analysis, cluster analysis, partial least squares discriminant analysis) and univariate analysis (including non-parametric test or parametric test, multiplicative analysis) using MetaboAnalyst 5.0 online analysis software to screen out the components with significant differences. Finally, pathway analysis was performed through the KEGG pathway database to identify the major metabolic pathways involved in the differential metabolites.
3. Result
3.1. Physicochemical indexes of three dairy products
Compared with camel milk, the pH,viable bacteria count and acetaldehyde content of sour camel milk and oat jujube sour camel milk after 24 h of fermentation and 24 h of refrigeration at 4 °C were as shown in Table 1.
Table 1.
Physicochemical indexes.
| Projects | pH | viable bacterial count (CFU/mL) | acetaldehyde content (μg/mL) |
|---|---|---|---|
| Camel Milk | 6.5 ± 0.4 | \ | \ |
| Sour Camel Milk | 4.35 ± 0.23 | (2.05 ± 0.23)lg | 12.10 ± 0.39 |
| Oat jujube Sour Camel Milk | 4.68 ± 0.5 | (10.7 ± 0.09)lg | 13.63 ± 0.55 |
3.2. Nutritional composition of three types of dairy products
Compared with camel milk, the acidity, lactose, fat, protein and total sugar contents of sour camel milk and oat jujube sour camel milk after 24 h of fermentation and 24 h of refrigeration at 4 °C were as shown in Table 2, which all met the requirements of national food inspection standards.
Table 2.
Comparison of nutrients (%).
| Projects | Acidity | Lactose | Fat | Protein | Total Sugar |
|---|---|---|---|---|---|
| Camel Milk | 20 ± 0.04°T | 4.7 ± 0.32 | 5.6 ± 0.38 | 3.8 ± 0.26 | 8.6 ± 0.14 |
| Oat jujube Sour Camel Milk | 143 ± 0.64°T | 3.9 ± 0.26 | 5.1 ± 0.26 | 4.1 ± 0.81 | 8.2 ± 0.07 |
| Sour Camel Milk | 143 ± 0.05°T | 4.3 ± 0.65 | 5.1 ± 0.57 | 3.3 ± 0.41 | 1.2 ± 0.12 |
| Inspection Standards | ≥77°T | \ | ≥3.1 | ≥2.9 | \ |
3.3. Metabolomics multivariate statistical analysis
3.3.1. Principal component analysis
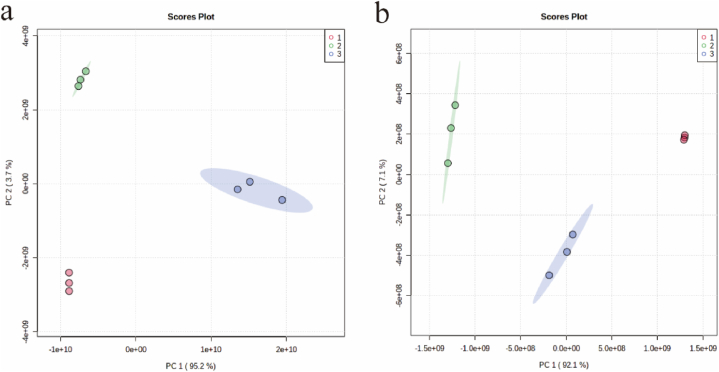
In this study, the metabolites of camel milk and its fermented products were analyzed by PCA analysis. The similarity of metabolic components between samples can be seen from the degree of aggregation and dispersion of each sample in the principal component analysis plot, where the closer the position is, the more similar it is, and vice versa. As shown in Fig. 1, samples from different groups showed aggregated status on the graph, indicating that the metabolites in each group were more similar, and the discrete status with large differences between groups indicated that the metabolic components were more different between groups, which provided a prerequisite for the subsequent screening of differential metabolites. From the PCA score plots (Fig. 1a and b), the PCA model had a good fit, and the samples were all within the 95 % confidence interval. The camel milk, sour camel milk and oat jujube sour camel milk samples were obviously clustered into three groups, which were basically effectively distinguished, indicating that there were obvious differences in the components between the groups, which could be identified by PCA identification, and the data were worthy of further study.
Fig. 1.
Similarity of metabolic components between samples (a); PCA score graph in positive ion mode (b); PCA score graph in negative ion mode. 1, camel milk; 2, sour camel milk; 3, oat jujube sour camel milk.
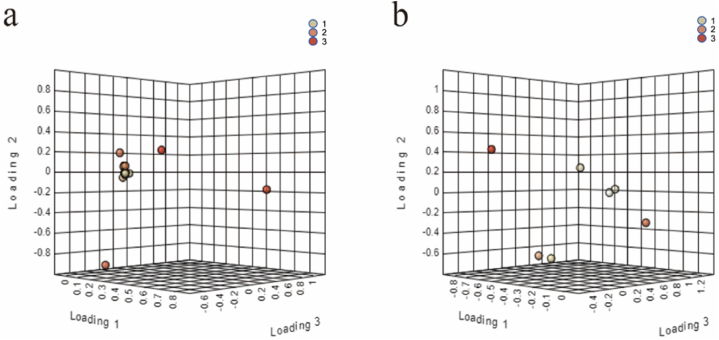
The load is an indication of the relationship between each variable and the principal component in the principal component analysis (Fig. 2), which mainly reflects the degree of influence of the metabolic component on the principal component, as shown in Fig. 2a and b. The more distant the variables from the origin, the greater the relative weight coefficient and the greater the influence on the principal component. Combined with the subsequent annotation information, it contributes to biomarker discovery.
Fig. 2.
Effect of metabolic components on principal components (a); PCA load map in positive ion mode (b); PCA load map in negative ion mode. 1, camel milk; 2, sour camel milk; 3, oat jujube sour camel milk.
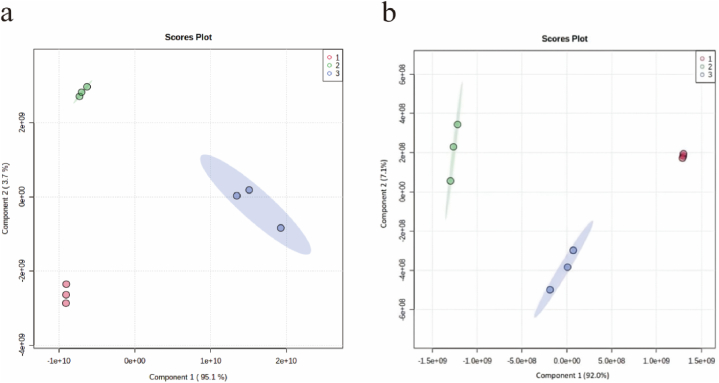
3.3.2. Partial least squares discriminant analysis
Although principal component analysis is effective in extracting primary information, it is insensitive to variables with low correlation, which can be addressed by partial least squares discriminant analysis. PLS-DA is a supervised discriminant analysis method for metabolomic data classification and regression, which can ignore random errors and make data analysis more focused and accurate [49,50]. To further differentiate the metabolites of camel milk, sour camel milk, and oat jujube sour camel milk, the PLS-DA model was used in this study to analyze the metabolic information of each of the three dairy products. The components of the three groups were predicted as shown in Fig. 3, and the PLS-DA scores of the positive ion (Fig. 3a) and negative ion (Fig. 3b) models were plotted, with the vertical coordinates representing the intra-group differences and the horizontal coordinates representing the inter-group differences, from which it can be seen that the inter-group differences were obvious and did not overlap. In addition, the R2Y of the discriminative combination model for different dairy products was 1.00, indicating that up to 100 % of the original data were retained. the predictive ability of the PLS-DA model Q2Y was above 0.95, and R2Y was greater than Q2Y, indicating that the developed model had good predictive ability.
Fig. 3.
Partial least squares discriminant analysis (a); PLS-DA score chart in positive ion mode (b); PLS-DA score chart in negative ion mode. 1, camel milk; 2, sour camel milk; 3, oat jujube sour camel milk.
3.4. Differential metabolite analysis
3.4.1. Classification of differential metabolites
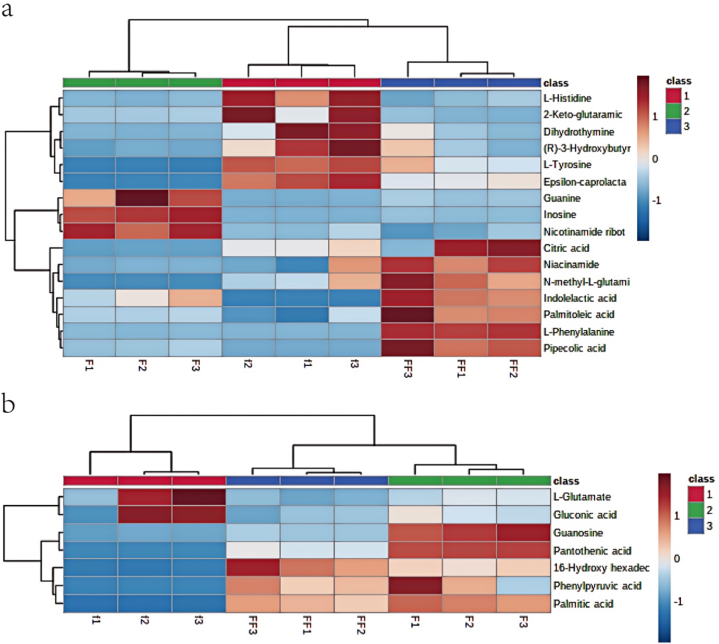
Hierarchical cluster analysis visualises the differences in metabolite content between camel milk, sour camel milk and oat jujube sour camel milk in the form of a heat map. In the heat map of the cluster analysis, the same row represents the same differential metabolite, and the same column represents the same camel milk sample. Different colours represent different levels of different metabolites. Red indicates high levels and blue indicates low levels [51]. The differential metabolites of camel milk, sour camel milk and oat jujube sour camel milk were analyzed in clusters and represented as heat maps. There is a clear colour distribution indicating distinct zones of high and low expression in the different camel milk samples, which can be used to differentiate the functional metabolites of camel milk.
As shown in Fig. 4, a comprehensive cluster analysis was conducted for the three dairy products (camel milk, sour camel milk and oat jujube sour camel milk). The distribution of the differential metabolites in the three samples is presented in the form of a heat map. The metabolites identified after clustering analysis were amino acids, nucleoside compounds, organic. acids and amides (Fig. 4a). The bioactive compounds in the positive ion mode included l-phenylalanine, l-tyrosine, l-histidine N-methyl-l-glutamic acid, hypoxanthine ribose, nicotinamide ribose, guanine, dihydrothymidine, citric acid, percarbonic acid, α-ketoglutaric acid, (R)-3-hydroxybutyric acid, indolyl lactate, palmitic acid, nicotinamide, and ε-caprolactam. Among them, ε -caprolactam, l-tyrosine, (R)-3-hydroxybutyric acid, dihydrothymidine, α-ketoglutaric acid and l-histidine were significantly higher in camel milk samples than in sour camel milk and oat jujube sour camel milk, with a clear distribution of high and low contents, (high metabolites (red) were distributed above and low metabolites (blue) were distributed below). Nicotinamide Ribose, inosine and guanine were significantly higher in the sour camel milk sample than in the camel milk and oat jujube sour camel milk, with the high metabolites (red) distributed in the middle and the low metabolites (blue) distributed on both sides. High alcoholic acid, l-phenylalanine, indole lactic acid, palmitic acid, N-methyl-l-glutamic acid, nicotinamide and citric acid were significantly higher in the oat jujube sour camel milk sample than in the camel milk and sour camel milk, high metabolites (red) were distributed at the bottom and low metabolites (blue) were distributed at the top (Fig. 4b). Bioactive compounds in the negative ion mode included l-glutamic acid, guanosine, phenylpyruvic acid, palmitic acid, gluconic acid, pantothenic acid and 16-hydroxyhexadecanoic acid. The levels of gluconic acid and l-glutamic acid were significantly higher in camel milk samples than in acidic camel milk, oat and date camel milk (high metabolites (red) at the top and low metabolites (blue) at the bottom); the levels of phenylpyruvic acid, palmitic acid and 16-hydroxyhexadecanoic acid were significantly higher in acidic camel milk and oat and date camel milk (high metabolites (red) at the bottom) and 16-hydroxyhexadecanoic acid at the bottom. Pantothenic acid and guanosine were significantly higher in acidic camel milk than in camel milk and oat-date acidic camel milk (high metabolites (red) at the bottom and low metabolites (blue) at the top).
Fig. 4.
The heat maps of cluster analysis of all differential metabolites of camel milk, sour camel milk and oat jujube sour camel milk (a); Heat map of differential component clustering in positive ion mode (b); Heat map of differential component clustering in negative ion mode 1, camel milk; 2, sour camel milk; 3, oat jujube sour camel milk.
Cluster analysis of differential metabolites accurately identifies those with significant differences among dairy products. By comparing these differences, the three dairy products were successfully distinguished. The heat map of metabolites reveals their types and contents, showcasing distinct color distributions. These differences in color distribution indicate functional diversity among the metabolites of the three dairy products.
3.4.2. Differential metabolite pathway analysis
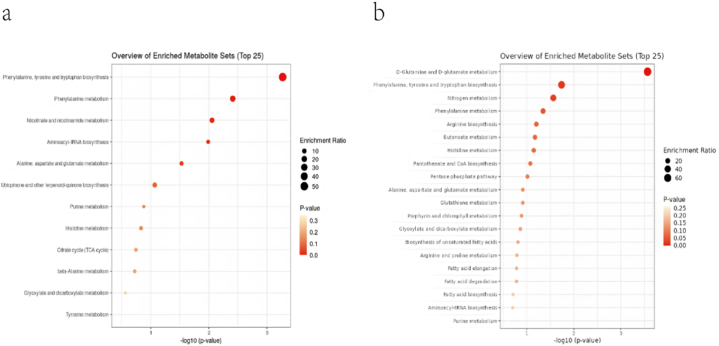
The components were compared with the KEGG pathway database to collate all pathways mapping to the partially differential components of the corresponding species. The metabolic pathways involved in the differential metabolites of camel milk, sour camel milk, and oat and red date sour camel milk are shown in (Fig. 5), and the identification results without categorical information were not involved in the statistics. As shown in (Fig. 5a), the metabolisms involved in the positive ion mode are phenylalanine, niacin and nicotinamide, purine (Fig. 5b), Metabolisms involved in the negative ion mode are phenylalanine, fatty acids, unsaturated fatty acid biosynthesis; butyric acid, glyoxylic acid and dicarboxylic acid, purine.
Fig. 5.
Metabolic pathways involved in different metabolites (a); Bar graph of metabolic pathway analysis in positive ion mode (b); Bar graph of metabolic pathway analysis in negative ion mode.
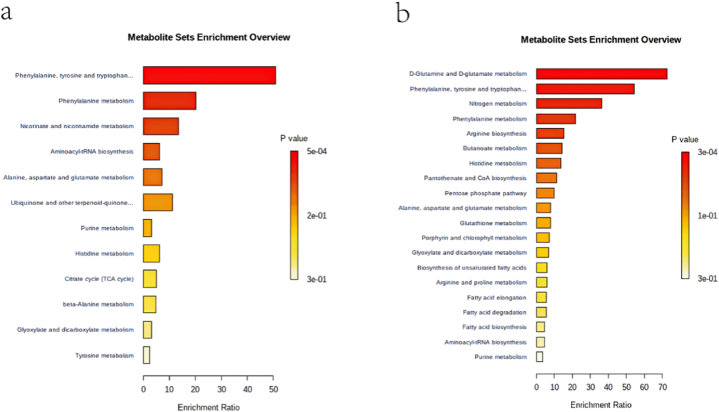
To obtain the close relationship between the metabolic pathways and the experimental conditions, enrichment analysis was performed on the metabolic pathways where the differential components were located. After obtaining the matching information of the differential components, we searched the corresponding pathway database and completed the metabolic pathway enrichment analysis and filtered the pathways containing the differential components by calculating the enrichment ratios and p-values to make bubble plots. As shown in (Fig. 6), the horizontal coordinate is the negative logarithm of the p-value, the vertical coordinate is the name of the metabolic pathway, and the color of the bubble is the size of the p-value, the smaller the p-value means the more significant enrichment; the size of the bubble is the size of the enrichment ratio, the larger the bubble means the larger the enrichment ratio, the more the number of metabolites, the more valuable the metabolic pathway is for research. As can be seen from (Fig. 6a), the metabolic pathways in which the metabolites are significantly involved in the positive ion mode are mainly the biosynthesis of phenylalanine, tyrosine, tryptophan and the metabolism of phenylalanine, nicotinic acid and nicotinamide, etc. The metabolites involved in the biosynthesis of phenylalanine, tyrosine and tryptophan are l-phenylalanine and l-tyrosine, the metabolite involved in the metabolism of phenylalanine is l-phenylalanine, and the metabolite involved in the metabolism of nicotinic acid and nicotinamide is nicotinamide. As can be seen from (Fig. 6b), the metabolic pathways in which metabolites are significantly different in the negative ion mode are mainly d-glutamine and d-glutamate and nitrogen metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, etc. The metabolite involved in d-glutamine and d-glutamate metabolism is l-glutamate, the metabolite involved in phenylalanine, tyrosine and tryptophan biosynthesis is phenylpyruvate, and the metabolite involved in nitrogen metabolism is guanosine.
Fig. 6.
Enrichment analysis of metabolic pathways of different components (a); Bubble diagram of metabolic pathway enrichment analysis in positive ion mode (b); Bubble diagram of metabolic pathway enrichment analysis in negative ion mode.
4. Discussion
Camel milk is widely known for being rich in vitamins, unsaturated fatty acids, lactoferrin, immunoglobulins and high levels of insulin, among other nutrients that are beneficial to human health [52]. In this study, fresh camel milk was fermented using laboratory screening of lactic acid bacteria and optimal fermentation conditions to prepare sour camel milk. In addition, 1 % glucose, 5 % jujube powder and 3 % oatmeal were added during the fermentation process to prepare oat jujube sour camel milk, so that camel milk has both nutritional value and many probiotic functions at the same time. Liquid chromatography-mass spectrometry (LC-MS) untargeted metabolomics was used to determine the differential metabolites of camel milk, fermentative bacterium TR1 fermented sour camel milk and oat red date sour camel milk. Among the differential metabolites, the levels of citric acid, nicotinamide, N-methyl-l-glutamic acid, indolyl lactate, palmitoleic acid, l-phenylalanine, and percocet were significantly higher in sour camel milk than in camel milk and oat and jujube sour camel milk. Among these substances, nicotinamide is a vitamin with potent anti-inflammatory properties [53]. Niacinamide deficiency can lead to pellagra, dermatitis, diarrhea, insomnia, headache, and other symptoms. A derivative of nicotinamide is coenzyme I, which can be used as a coenzyme for many kinds of dehydrogenases in the body. In addition, it has three important biological functions: as an energy donor for DNA ligase in bacterial cells, involved in ADP-ribosylation modification of proteins, and involved in deacetylation modification of histones. This suggests that yogurt can be a good supplement for niacinamide. Currently, niacinamide is mainly used in pharmaceuticals and cosmeceuticals, and studies have confirmed that the use of niacinamide as an essential nutrient will benefit the health of the whole body and skin [54,55].A study by De Souza et al. [56] showed that palmitoleic acid plays an important role as a non-pharmacological treatment in the treatment of diabetes mellitus and liver dysfunction. This is in line with the study of Hussain et al. [57], where camel milk significantly reduced blood glucose, along with HbA1c (p < 0.01), aspartate aminotransferase (AST), alanine aminotransferase (ALT) (p < 0.1), triglyceride (TG), and cholesterol (p < 0.1), which could be used as a suitable alternative for the treatment of diabetes.The higher levels of palmitoleic acid in sour camel milk compared to camel milk predicts a role for sour camel milk in diabetes and liver dysfunction. l-phenylalanine is an essential amino acid, slightly bitter in taste, and belongs to the aromatic amino acids. In addition, lysine, methionine, leucine and histidine, which are essential for human body, are all in the form of derivatives in the different fractions, and most of the essential amino acid derivatives are significantly higher in sour camel milk than camel milk, so sour camel milk can provide essential amino acids for human body more effectively. The contents of guanine, inosine and nicotinamide riboside in oat jujube sour camel milk were significantly higher than those in the other two groups. Inosine has been reported to have anti-inflammatory and immunomodulatory properties [58,59], predicting that oat jujube sour camel milk could play an important role in anti-inflammation and immunomodulation. In addition, inosine is a dietary supplement widely used in the treatment of many central nervous system disorders, and recent experimental studies on inosine have revealed its potential to promote peripheral neuroprotection after sciatic nerve injury, and inosine is highly promising in attenuating diabetic peripheral neuropathy (DPN) in rats, possibly as a potential signaling pathway [60]. Recent findings suggest that nicotinamide riboside (NR) and its phosphorylated form nicotinamide mononucleotide (NMN) are also effective in inducing NAD biosynthesis, complementing other recognized NAD precursors, such as NMN or NR, which have been shown to reduce neuronal loss after brain injury in mice by increasing intracellular NAD content, and have therapeutic utility in clinical studies [61]. Therefore, oat jujube sour camel milk is expected to perform its probiotic function in the treatment of neurological diseases.
The KEGG database was used to annotate the differential metabolic pathways in different ionic modes, with 12 major metabolic pathways involved in positive ionic mode and 20 major metabolic pathways involved in negative ionic mode. The results showed that the camel milk, sour camel milk and oat jujube sour camel milk changed significantly and differed in the metabolic pathways of amino acids, nitrogen, organic acids and purines, and the nutritional quality and probiotic functions of camel milk were enhanced by lactic acid bacteria fermentation and the addition of oat and jujube during fermentation.
5. Conclusion
In this study, we compared the differential fractions of sour camel milk and oat jujube sour camel milk prepared by lactic acid bacterial agent TR1 with camel milk by liquid chromatography-mass spectrometry (LC-MS) non-targeted metabolomics, and found that the addition of oat and jujube during fermentation and fermentation of lactic acid bacterial agent TR1 significantly affected the metabolic pathways of amino acid metabolism, nitrogen metabolism, organic acid metabolism, and purine metabolism of the three products by affecting The content of amino acids, nucleosides, organic acids and amides of the three products was significantly increased in sour camel milk compared with camel milk, and the content of beneficial components such as niacinamide, l-phenylalanine and palmitic acid was significantly increased in oat jujube sour camel milk compared with camel milk, and the content of beneficial components such as guanine, inosine and niacinamide riboside was significantly increased in oat jujube sour camel milk compared with camel milk, which showed that sour camel milk and oat jujube sour camel milk improved the nutritional quality and probiotic function of camel milk The results showed that camel milk and oat jujube sour camel milk improved the nutritional quality and probiotic function of camel milk, and provided a theoretical basis for the development of processing technology and products related to camel milk and fermented camel milk.
Funding
This research was funded by three grant programs:1. Team funding for "Germplasm Innovation and Molecular Breeding of Forage Crops and Useful Microbiology" (TD202103); 2. Youth Fund of College of Life Sciences, Inner Mongolia Agricultural University (QN202103); 3. Inner Mongolia Natural Science Foundation Program (2023MS03047)
Data availability statement
The data presented in this study are available on request from the corresponding author.
Institutional Review Board Statement.
The study did not involve humans or animals.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Qingwen Guo: Writing – review & editing, Writing – original draft, Software, Methodology, Investigation, Data curation, Conceptualization. Qigeqi Dong: Writing – review & editing, Project administration, Funding acquisition, Formal analysis. Weisheng Xu: Visualization, Software, Data curation. Heping Zhang: Methodology, Formal analysis, Conceptualization. Xiangyu Zhao: Validation, Formal analysis. Wanxiong He: Software, Investigation. Yuxing He: Investigation, Formal analysis. Guofen Zhao: Writing – review & editing, Supervision, Project administration, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Yaqoob M, Nawaz H. Potential of Pakistani camel for dairy and other uses [J]. Anim. Sci. J., 78(5): 467-475. doi: 10.1111/j.1740-0929.2007.00464.x.
- 2.Rahmeh R., Akbar A., Kishk M., AL-Onaizi T., AL-Azmi A., AL-Shatti A., Shajan A., AL-Mutairi S., Akbar B. Distribution and antimicrobial activity of lactic acid bacteria from raw camel milk. New Microbes New Infect. 2019;30 doi: 10.1016/j.nmni.2019.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benmeziane-Derradji F. Evaluation of camel milk: gross composition-a scientific overview. Trop. Anim. Health Prod. 2021;53(2):308. doi: 10.1007/s11250-021-02689-0. [DOI] [PubMed] [Google Scholar]
- 4.Hamed A.I., Ben S.A.I.D.R., Kontek B., AL-Ayed A.S., Kowalczyk M., Moldoch J., Oleszek W., Stochmal A., Olas B. Electrospray ionization mass spectrometry characterization of ubiquitous minor lipids and oligosaccharides in milk of the camel (Camelus dromedarius) and their inhibition of oxidative stress in human plasma. J. Dairy Sci. 2020;103(1):72–86. doi: 10.3168/jds.2019-16710. [DOI] [PubMed] [Google Scholar]
- 5.Oselu S., Ebere R., Arimi JM. Camels, camel milk, and camel milk product situation in Kenya in relation to the world. Int. J. Food Sci. 2022;2022:1237423. doi: 10.1155/2022/1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amrouche T., Mounier J., Pawtowski A., Thomas F., Picot A. Microbiota associated with dromedary camel milk from Algerian sahara. Curr. Microbiol. 2020;77(1):24–31. doi: 10.1007/s00284-019-01788-4. [DOI] [PubMed] [Google Scholar]
- 7.Arain M.A., Khaskheli G.B., Shah A.H., Marghazani I.B., Barham G.S., Shah Q.A., Khand F.M., Buzdar J.A., Soomro F., Fazlani S.A. Nutritional significance and promising therapeutic/medicinal application of camel milk as a functional food in human and animals: a comprehensive review. Anim. Biotechnol. 2023;34(6):1988–2005. doi: 10.1080/10495398.2022.2059490. [DOI] [PubMed] [Google Scholar]
- 8.Ayyash M., AL-Nuaimi A.K., AL-Mahadin S., Liu S.Q. In vitro investigation of anticancer and ACE-inhibiting activity, alpha-amylase and alpha-glucosidase inhibition, and antioxidant activity of camel milk fermented with camel milk probiotic: a comparative study with fermented bovine milk. Food Chem. 2018;239:588–597. doi: 10.1016/j.foodchem.2017.06.149. [DOI] [PubMed] [Google Scholar]
- 9.Althwab S.A., Alamro S.A., AL Abdulmonem W., Allemailem K.S., Alarifi S.A., Hamad E.M. Fermented camel milk enriched with plant sterols improves lipid profile and atherogenic index in rats fed high -fat and -cholesterol diets. Heliyon. 2022;8(10) doi: 10.1016/j.heliyon.2022.e10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakry I.A., Yang L., Farag M.A., Korma S.A., Khalifa I., Cacciotti I., Ziedan N.I., Jin J., Jin Q., Wei W., Wang X. A comprehensive review of the composition, nutritional value, and functional properties of camel milk fat. Foods. 2021;10(9) doi: 10.3390/foods10092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao W., He Y., Liu W. Diversity analysis of bacterial and function prediction in hurunge from Mongolia. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.835123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D., Li X.Y., Zhao X., Qin Y.S., Zhang X.X., Li J., Wang J.M., Wang C.F. Proteomics and microstructure profiling of goat milk protein after homogenization. J. Dairy Sci. 2019;102(5):3839–3850. doi: 10.3168/jds.2018-15363. [DOI] [PubMed] [Google Scholar]
- 13.He J., Xiao Y., Orgoldol K., Ming L., Yi L., Ji R. Effects of geographic region on the composition of bactrian camel milk in Mongolia. Animals (Basel) 2019;9(11) doi: 10.3390/ani9110890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ider S., Belguesmia Y., Coucheney F., Kihal M., Drider D. Impact of seasonality and environmental conditions on yeast diversity from camel's milk collected in Algeria. Arch. Microbiol. 2019;201(3):399–407. doi: 10.1007/s00203-019-01626-y. [DOI] [PubMed] [Google Scholar]
- 15.Shaban A.M., Raslan M., Qahl S.H., Elsayed K., Abdelhameed M.S., Oyouni A.A.A., AL-Amer O.M., Hammouda O., EL-Magd M.A. Ameliorative effects of camel milk and its exosomes on diabetic nephropathy in rats. Membranes. 2022;12(11) doi: 10.3390/membranes12111060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchard J., Valookaran A.F., Aloud B.M., Raj P., Malunga L.N., Thandapilly S.J., Netticadan T. Impact of oats in the prevention/management of hypertension. Food Chem. 2022;381 doi: 10.1016/j.foodchem.2022.132198. [DOI] [PubMed] [Google Scholar]
- 17.Li X., Cai X., Ma X., Jing L., Gu J., Bao L., Li J., Xu M., Zhang Z., Li Y. Short- and long-term effects of wholegrain oat intake on weight management and glucolipid metabolism in overweight type-2 diabetics: a randomized Control trial. Nutrients. 2016;8(9) doi: 10.3390/nu8090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fant P., Ramin M., Huhtanen P. Replacement of barley with oats and dehulled oats: effects on milk production, enteric methane emissions, and energy utilization in dairy cows fed a grass silage-based diet. J. Dairy Sci. 2021;104(12):12540–12552. doi: 10.3168/jds.2021-20409. [DOI] [PubMed] [Google Scholar]
- 19.Musa-Veloso K., Noori D., Venditti C., Poon T., Johnson J., Harkness L.S., O'Shea M., Chu Y. A systematic review and meta-analysis of randomized controlled trials on the effects of oats and oat processing on postprandial blood glucose and insulin responses. J. Nutr. 2021;151(2):341–351. doi: 10.1093/jn/nxaa349. [DOI] [PubMed] [Google Scholar]
- 20.Zhao M., Zhang H., Wang J., Shan D., Xu Q. Serum metabolomics analysis of the intervention effect of whole grain oats on insulin resistance induced by high-fat diet in rats. Food Res. Int. 2020;135 doi: 10.1016/j.foodres.2020.109297. [DOI] [PubMed] [Google Scholar]
- 21.Ma L., Luo Z., Huang Y., Li Y., Guan J., Zhou T., DU Z., Yong K., Yao X., Shen L., Yu S., Zhong Z., Hu Y., Peng G., Shi X., Cao S. Modulating gut microbiota and metabolites with dietary fiber oat beta-glucan interventions to improve growth performance and intestinal function in weaned rabbits. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1074036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Cordero J., Sierra-Cinos J.L., Seguido M.A., Gonzalez-Ramila S., Mateos R., Bravo-Clemente L., Sarria B. Regular consumption of green coffee phenol, oat beta-glucan and green coffee phenol/oat beta-glucan supplements does not change body composition in subjects with overweight and obesity. Foods. 2022;11(5) doi: 10.3390/foods11050679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang K., Zhang S., Guan X., Li C., Li S., Liu Y., Shi J. Effect of the oat beta-glucan on the development of functional quinoa (Chenopodium quinoa wild) milk. Food Chem. 2021;349 doi: 10.1016/j.foodchem.2021.129201. [DOI] [PubMed] [Google Scholar]
- 24.Wolever T.M.S., Rahn M., Dioum E.H., Jenkins A.L., Ezatagha A., Campbell J.E., Chu Y. Effect of oat beta-glucan on affective and physical feeling states in healthy adults: evidence for reduced headache, fatigue, anxiety and limb/joint pains. Nutrients. 2021;13(5) doi: 10.3390/nu13051534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva F.A., Queiroga R., DE Souza E.L., Voss G.B., Borges G., Lima M.D.S., Pintado M.M.E., Vasconcelos M. Incorporation of phenolic-rich ingredients from integral valorization of Isabel grape improves the nutritional, functional and sensory characteristics of probiotic goat milk yogurt. Food Chem. 2022;369 doi: 10.1016/j.foodchem.2021.130957. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y., Bao T., Mo J., Ni J., Chen W. Research advances in bioactive components and health benefits of jujube (Ziziphus jujuba Mill.) fruit. J. Zhejiang Univ. - Sci. B. 2021;22(6):431–449. doi: 10.1631/jzus.B2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q.H., Wu C.S., Wang M. The jujube (Ziziphus jujuba Mill.) fruit: a review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 2013;61(14):3351–3363. doi: 10.1021/jf4007032. [DOI] [PubMed] [Google Scholar]
- 28.Yekta M., Ansari S. Jujube mucilage as a potential stabilizer in stirred yogurt: improvements in the physiochemical, rheological, and sensorial properties. Food Sci. Nutr. 2019;7(11):3709–3721. doi: 10.1002/fsn3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng C., Wang B., Zhao A., Wei L., Shao Y., Wang Y., Cao B., Zhang F. Quality characteristics and antioxidant activities of goat milk yogurt with added jujube pulp. Food Chem. 2019;277:238–245. doi: 10.1016/j.foodchem.2018.10.104. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X H. Preparation of Lyophilized Lactic Acid Bacteria for Camel Milkfermentation [M.S. Thesis]. Inner Mongolia Agricultural University.(In Chinese).
- 31.Aljutaily T, Rehan M, Moustafa M M A, Barakat H. Effect of intermittent fasting, probiotic-fermented camel milk, and Pr obiotic-fermented camel milk incorporating sukkari date on diet-induce d obesity in rats [J]. Fermentation, 8(11): 619. doi: 10.3390/fermentation8110619.
- 32.Schultheiss U.T., Sekula P. The promise of metabolomics in decelerating CKD progression in children. Clin. J. Am. Soc. Nephrol. 2021;16(8):1152–1154. doi: 10.2215/CJN.07400521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu H., Zhang W., Yun Y., Chen W., Zhong Q., Hu Y., Chen H., Chen W. Metabolomics study on revealing the inhibition and metabolic dysregulation in Pseudomonas fluorescens induced by 3-carene. Food Chem. 2020;329 doi: 10.1016/j.foodchem.2020.127220. [DOI] [PubMed] [Google Scholar]
- 34.Gika H., Virgiliou C., Theodoridis G., Plumb R.S., Wilson I.D. Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): the state of the art. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2019;1117:136–147. doi: 10.1016/j.jchromb.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Arapitsas P., Mattivi F. LC-MS untargeted protocol for the analysis of wine. Methods Mol. Biol. 2018;1738:225–235. doi: 10.1007/978-1-4939-7643-0_16. [DOI] [PubMed] [Google Scholar]
- 36.Ji L., Chen S., Gu G., Wang W., Ren J., Xu F., Li F., Wu J., Yang D., Zheng Y. Discovery of potential biomarkers for human atherosclerotic abdominal aortic aneurysm through untargeted metabolomics and transcriptomics. J. Zhejiang Univ. - Sci. B. 2021;22(9):733–745. doi: 10.1631/jzus.B2000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo C Y, Yao X Z, Chen J, Lv L T. Analysis of substance differences before and after fermentation of black tea infusion by eurotium cristatum based on non-targeted metabonomics [J]. Food Science and Technology, 46(5), 246-253. (In Chinese).
- 38.Mckee L.S. Measuring enzyme kinetics of glycoside hydrolases using the 3,5-dinitrosalicylic acid assay. Methods Mol. Biol. 2017;1588:27–36. doi: 10.1007/978-1-4939-6899-2_3. [DOI] [PubMed] [Google Scholar]
- 39.Grintzalis K., Georgiou C.D., Schneider Y.J. An accurate and sensitive Coomassie Brilliant Blue G-250-based assay for protein determination. Anal. Biochem. 2015;480:28–30. doi: 10.1016/j.ab.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 40.Kleyn D.H., Lynch J.M., Barbano D.M., Bloom M.J., Mitchell M.W. Determination of fat in raw and processed milks by the Gerber method: collaborative study. J. AOAC Int. 2001;84(5):1499–1508. [PubMed] [Google Scholar]
- 41.Portnoy M., Barbano D.M. Lactose: use, measurement, and expression of results. J. Dairy Sci. 2021;104(7):8314–8325. doi: 10.3168/jds.2020-18706. [DOI] [PubMed] [Google Scholar]
- 42.Escuder-Vieco D., Vazquez-Roman S., Sanchez-Pallas J., Ureta-Velasco N., Mosqueda-Pena R., Pallas-Alonso C.R. Determination of acidity in donor milk. J Hum Lact. 2016;32(4):NP73–N75. doi: 10.1177/0890334415591338. [DOI] [PubMed] [Google Scholar]
- 43.Abdelhafez O.H., Othman E.M., Fahim J.R., Desoukey S.Y., Pimentel-Elardo S.M., Nodwell J.R., Schirmeister T., Tawfike A., Abdelmohsen U.R. Metabolomics analysis and biological investigation of three Malvaceae plants. Phytochem. Anal. 2020;31(2):204–214. doi: 10.1002/pca.2883. [DOI] [PubMed] [Google Scholar]
- 44.Monnerat G., Seara F.A.C., Evaristo J.A.M., Carneiro G., Evaristo G.P.C., Domont G., Nascimento J.H.M., Mill J.G., Nogueira F.C.S., Campos D.E., Carvalho A.C. Aging-related compensated hypogonadism: role of metabolomic analysis in physiopathological and therapeutic evaluation. J. Steroid Biochem. Mol. Biol. 2018;183:39–50. doi: 10.1016/j.jsbmb.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 46.Kim T.J., Hyeon H., Park N.I., Yi T.G., Lim S.H., Park S.Y., Ha S.H., Kim J.K. A high-throughput platform for interpretation of metabolite profile data from pepper (Capsicum) fruits of 13 phenotypes associated with different fruit maturity states. Food Chem. 2020;331 doi: 10.1016/j.foodchem.2020.127286. [DOI] [PubMed] [Google Scholar]
- 47.Farag M A, Sallam I E, Fekry M I, Zaghloul S S, EL-Dine R S. Metabolite profiling of three Opuntia ficus-indica fruit cultivars usi ng UPLC-QTOF-MS in relation to their antioxidant potential [J]. Food Biosci., 36: 100673. doi: 10.1016/j.fbio.2020.100673.
- 48.Silva L.N., Rigo G.V., Silva D.B., Carollo C.A., Trentin D.S., Silva M.V., Tasca T., Macedo A.J. Hydrolyzable tannins from Poincianella (Caesalpinia) microphylla fruits: metabolite profiling and anti-Trichomonas vaginalis activity. Food Res. Int. 2020;134 doi: 10.1016/j.foodres.2020.109236. [DOI] [PubMed] [Google Scholar]
- 49.Valderrama L., Valderrama P., Carasek E. A semi-quantitative model through PLS-DA in the evaluation of carbendazim in grape juices. Food Chem. 2022;368 doi: 10.1016/j.foodchem.2021.130742. [DOI] [PubMed] [Google Scholar]
- 50.Mansuri S M, Chakraborty S K, Mahanti N K, Pandiselvam R. Effect of germ orientation during Vis-NIR hyperspectral imaging for th e detection of fungal contamination in maize kernel using PLS-DA, ANN and 1D-CNN modelling [J]. Food Control, 139: 109077. doi: 10.1016/j.foodcont.2022.109077.
- 51.Zhang Q, Li B, Chen Q, Su Y, Wang R, Liu Z, Chen G. Non-targeted metabolomic analysis of the variations in the metabolites of two genotypes of Glycyrrhiza uralensis Fisch. under drought stress [J]. Ind. Crop. Prod., 176: 114402. doi: 10.1016/j.indcrop.2021.114402.
- 52.Aqib A I, Fakhar-E-Alam Kulyar M, Ashfaq K, Bhutta Z A, Shoaib M, Ahmed R. Camel milk insuline: pathophysiological and molecular repository [J]. Trends Food Sci. Technol., 88: 497-504. doi: 10.1016/j.tifs.2019.04.009.
- 53.Boo Y.C. Mechanistic basis and clinical evidence for the applications of nicotinamide (niacinamide) to Control skin aging and pigmentation. Antioxidants. 2021;10(8) doi: 10.3390/antiox10081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang H., Park Y.K., Lee J.Y. Nicotinamide riboside, an NAD(+) precursor, attenuates inflammation and oxidative stress by activating sirtuin 1 in alcohol-stimulated macrophages. Lab. Invest. 2021;101(9):1225–1237. doi: 10.1038/s41374-021-00599-1. [DOI] [PubMed] [Google Scholar]
- 55.Tang Y., Liu X., Feng C., Zhou Z., Liu S. Nicotinamide phosphoribosyltransferase (Nampt) of hybrid crucian carp protects intestinal barrier and enhances host immune defense against bacterial infection. Dev. Comp. Immunol. 2022;128 doi: 10.1016/j.dci.2021.104314. [DOI] [PubMed] [Google Scholar]
- 56.DE Souza C.O., Teixeira A.A.S., Biondo L.A., Lima Junior E.A., Batatinha H.A.P., Rosa Neto J.C. Palmitoleic acid improves metabolic functions in fatty liver by PPARalpha-dependent AMPK activation. J. Cell. Physiol. 2017;232(8):2168–2177. doi: 10.1002/jcp.25715. [DOI] [PubMed] [Google Scholar]
- 57.Hussain H., Wattoo F.H., Wattoo M.H.S., Gulfraz M., Masud T., Shah I., Ali S., Alavi S.E. Camel milk as an alternative treatment regimen for diabetes therapy. Food Sci. Nutr. 2021;9(3):1347–1356. doi: 10.1002/fsn3.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C., Ma Y., Wang D., Shan Y., Song X., Hu H., Ren X., Ma X., Cui J., Ma Y. Integrated microbiology and metabolomics analysis reveal plastic mulch film residue affects soil microorganisms and their metabolic functions. J. Hazard Mater. 2022;423(Pt B) doi: 10.1016/j.jhazmat.2021.127258. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y., Wang X., Zhu L., Tu Y., Chen W., Gong L., Pan T., Lin H., Lin J., Sun H., Ge Y., Wei L., Guo Y., Lu C., Chen Y., Xu L. Lactobacillus rhamnosus GG combined with inosine ameliorates alcohol-induced liver injury through regulation of intestinal barrier and Treg/Th1 cells. Toxicol. Appl. Pharmacol. 2022;439 doi: 10.1016/j.taap.2022.115923. [DOI] [PubMed] [Google Scholar]
- 60.Abdelkader N.F., Ibrahim S.M., Moustafa P.E., Elbaset M.A. Inosine mitigated diabetic peripheral neuropathy via modulating GLO1/AGEs/RAGE/NF-kappaB/Nrf2 and TGF-beta/PKC/TRPV1 signaling pathways. Biomed. Pharmacother. 2022;145 doi: 10.1016/j.biopha.2021.112395. [DOI] [PubMed] [Google Scholar]
- 61.Redeuil K., Vulcano J., Prencipe F.P., Benet S., Campos-Gimenez E., Meschiari M. First quantification of nicotinamide riboside with B(3) vitamers and coenzymes secreted in human milk by liquid chromatography-tandem-mass spectrometry. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2019;1110–1111:74–80. doi: 10.1016/j.jchromb.2019.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Institutional Review Board Statement.
The study did not involve humans or animals.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.