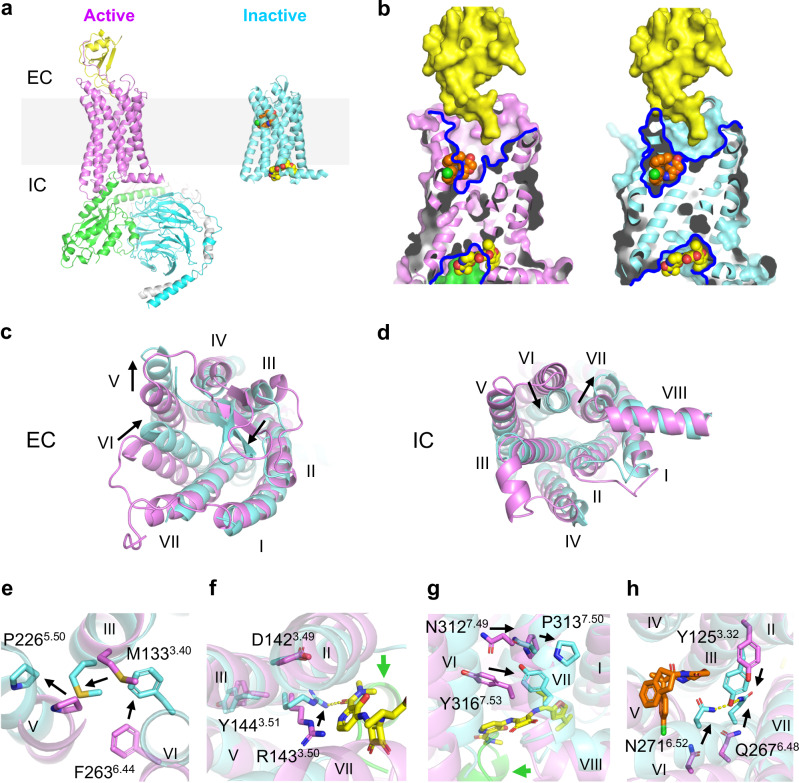
Fig. 5. Different mechanisms of allosteric antagonism of CCR6 by OXM and SQA.
a Overall structures of the active CCR6 (magenta, left, PDB ID 6WWZ) in complex with its cognate chemokine ligand CCL20 (yellow) and a heterotrimeric G protein coloured by subunit (Gαo, green, Gβ, cyan, and Gγ, light grey), and the inactive CCR6 (aquamarine, right) in complex with SQA1 (yellow carbon spheres) and OXM1 (orange carbon spheres). The light grey box highlights the location of cell membranes. b Overlays of CCL20, OXM1, and SQA1 bound structures onto the active (left) and the inactive (right) CCR6 structures. Structures are presented in the same colour codes as in a. Blue lines depict pockets revealed by the structures. c–d, Structural overlays of the active (magenta) and inactive (aquamarine) CCR6 structures from c EC and d IC views. e–h, Conformational changes of conserved functional motifs, e P2265.50-M1333.40-F2636.44, f D1423.49-R1433.50-Y1443.51, g N3127.49-P3137.50-x-x-Y3167.53, and h Y1253.32-Q2676.48-N2716.52 revealed by structural overlay of active (magenta) and inactive (aquamarine) CCR6. In c–h significant TM movements and side chain conformational changes from active to inactive states are highlighted by black arrows.