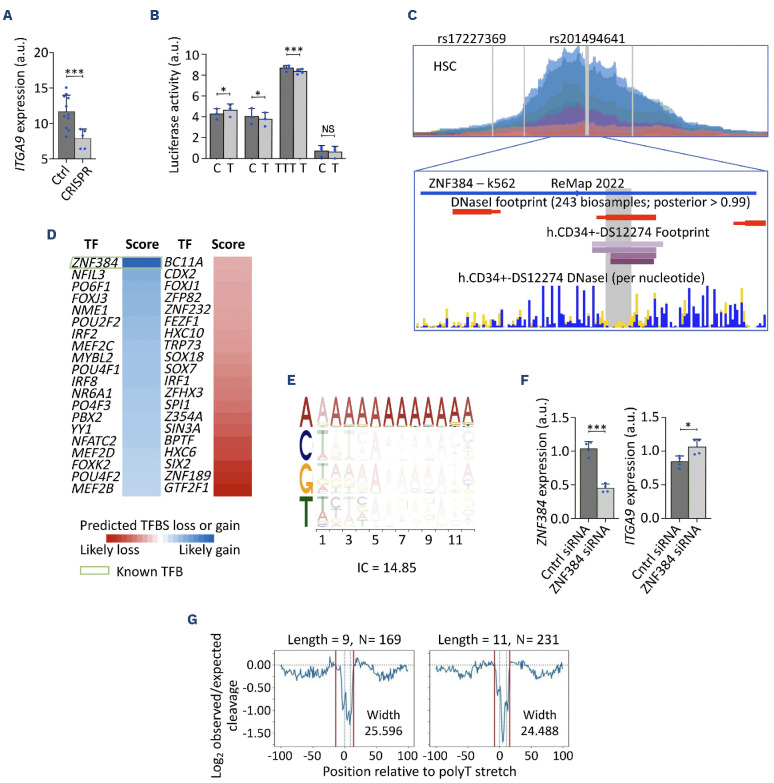
Figure 2.
ZNF384 preferential binding to rs201494641-T repressing ITGA9. (A) ITGA9 expression in HEL cells subjected to CRISPR/Cas9 editing with a non-targeting single-guide RNA (sgRNA) pair control (Ctrl, 60-bp cut, 11 biological replicates), or an sgRNA pair designed to delete the region harboring the 4 putative causal variants at 3p22 (CRISPR, 486-bp cut) (Online Supplementary Table S2); one-sided t test, ***P≤0.001. (B) Luciferase activities of the 4 candidate causal variants in HEL cells (4 biological replicates). Data normalized to empty vector control; one-sided t test, ***P≤0.001, *P≤0.05, NS: not significant. (C) Upper panel: chromatin accessibility (ATAC-sequencing signal intensity) across different blood cell types (colors as in Figure 1B) in the approximately 1,300-bp wide region in ITGA9 intron 3; lower panel: DNAse I footprint in primary CD34+ cells and chromatin immunoprecipitation sequencing (CHIP-seq) signals in K562 cells from ReMap in the rs201494641-harboring region. (D) ZNF384 gain of binding to rs201494641-T from the Fabian-variant database. (E) JASPAR detailed Transcription Factor Flexible Model TFFM0157.1 showing the ZNF384 consensus motif. (F) ZNF384 and ITGA9 expression in HEL cells transfected with Ctrl or ZNF384 small interfering RNA (siRNA) (4 biological replicates); one-sided t test, ***P≤0.001, *P≤0.05. (G) ENCODE DNAse I consensus footprint data for the ZNF384 motif in primary CD34+ cells, showing mean cleavage ratios in a 200-bp window centered on the core poly-T motif (dotted lines), for 2 different repeat lengths. In both cases the DNA binding footprint (solid lines) extends beyond the core poly-T stretch, suggesting that ZNF384 directly binds flanking sequences on either side of the repeat sequence. a.u.: arbitrary units; mRNA: messenger RNA; HSC: hematopoietic stem cells.