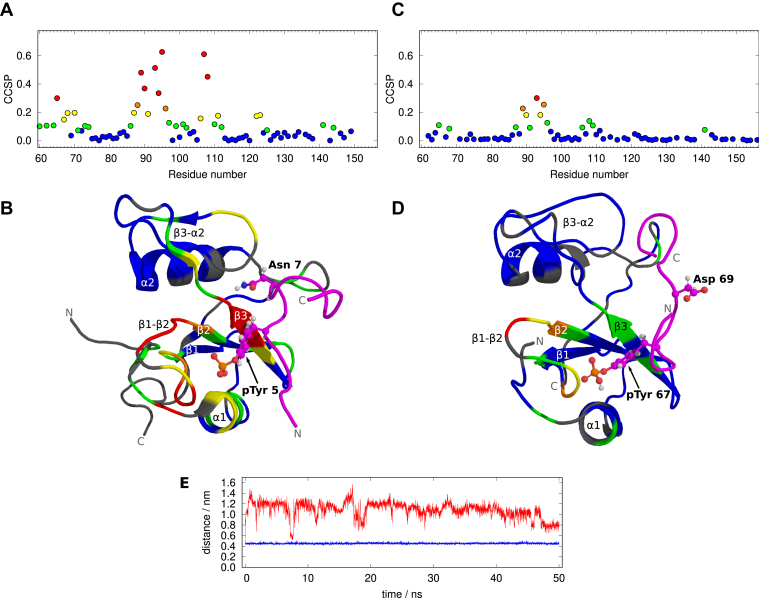
Figure 8.
Comparison of Grb2-SH2 in complexes with phosphoTyr-Val-Asn adn with MAP2c phosphorylated at Tyr67.A, CCSP of Grb2-SH2 in complex with phosphoTyr-Val-Asn (75). B, structure of Grb2-SH2 in complex with a phosphopeptide containing a pYVN motif (PDB ID 1QG1) (76), colored according to the CCSP plotted in panel A. Grb2-SH2 residues with missing CCSP values are shown in gray, the backbone trace of the phosphopeptide is shown in magenta. C, CCSP of Grb2-SH2 bound to rat MAP2c. The displayed CCSP values are calculated from the estimated KD for Grb2-SH2 fully saturated by Fyn-phosphorylated MAP2c. D, the lowest-energy structure of the molecular dynamics trajectory simulated for Grb2-SH2 in complex with Fyn-phosphorylated MAP2c, colored according to the CCSP plotted in panel C. Grb2-SH2 residues with missing CCSP values are shown in gray, the backbone trace of Fyn-phosphorylated MAP2c is shown in magenta. Residues pTyr5 and Asn7 of the pYVN motif and pTyr67 and Asp69 of Fyn-phosphorylated MAP2c are displayed as sticks and balls in panels B and D. E, distances between Cγ of the phosphopeptide Asn7/MAP2c Asp69 and carbonyl carbon of Grb2-SH2 Lys109 monitored during molecular dynamics simulation of Grb2-SH2 with phosphopeptides containing the pYVN sequence (blue) and the sequence derived from Fyn-phosphorylated MAP2c (red).