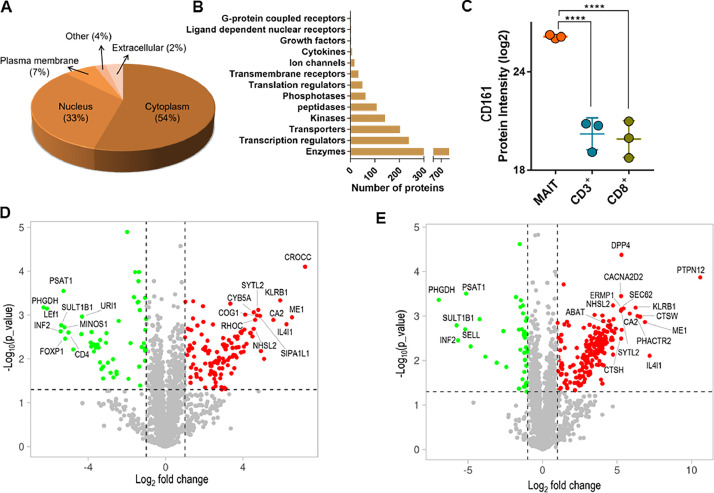
Fig. 3.
Gene ontology and differential expression analysis of normalized protein intensity data. A. Distribution of proteins in different subcellular compartments is given as a percentage of all proteins selected for differential expression analysis. B. Distribution of proteins selected for differential expression analysis across different functional groups (Qiagen, IPA). C. Expression of CD161 in three cell subsets as based on DDA-MS data (****q < 0.0001, multiple t-test with false discovery determination by two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli) D. Volcano plots labelling the top 20 differentially expressed proteins in MAIT cells compared to non-MAIT CD3+ T cells E. Volcano plots labelling the top 20 differentially expressed proteins in MAIT cells compared to non-MAIT CD8+ T cells.