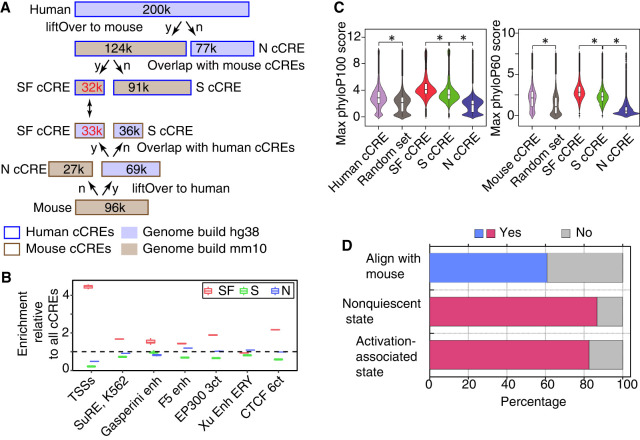
Figure 5.
Evolutionary and epigenetic comparisons of cCREs. (A) Workflow to partition blood cell cCREs in human and mouse into three evolutionary categories. (N) Nonconserved, (S) conserved in sequence but not inferred function, (SF) conserved in both sequence and inferred function as a cCRE, (y) yes, and (n) no. (B) Enrichment of SF-conserved human cCREs for TSSs. The number of elements in seven sets of function-related DNA intervals that overlap with the 32,422 SF human cCREs was determined, along with the number that overlap with three subsets (32,422 each) randomly selected from the full set of 200,342 human cCREs. The ratio of the number of function-related elements overlapping SF-cCREs to the number overlapping a randomly chosen subset of all cCREs gave the estimate of enrichment plotted in the graph. The mean for the three determinations of enrichment is indicated by the horizontal line for each set. Results are also shown for a similar analysis for the S and N cCREs. (C) Distribution of phyloP scores for three evolutionary categories of cCREs in human and mouse. The maximum phyloP score for each genomic interval was used to represent the score for each cCRE, using genome sequence alignments of 100 species with human as the reference (phyloP100) and alignments of 60 species with mouse as the reference (phyloP60). The distribution of phyloP scores for each group are displayed as a violin plot. All 10 random sets had distributions similar to the one shown. The asterisk (*) over brackets indicates comparison for which the P-value for Welch's t-test is <2.2 × 10−16. (D) Proportion of human genomic elements active in a massively parallel reporter assay (MPRA) that aligns with mouse or is in a state reflecting dynamic chromatin; 57,061 genomic elements found to be active in a lentivirus MPRA that tested a close to comprehensive set of predicted regulatory elements in K562 cells (Agarwal et al. 2023) were assessed for their ability to align with the mouse genome (blue bar) or whether the IDEAS epigenetic state assigned in K562 cells was not quiescent or was in a set of states associated with gene activation (magenta bars). The results are plotted as percentages of the total number of MPRA-active elements.