Abstract
Objectives
Escherichia coli can cause infections in the urinary tract and in normally sterile body sites leading to invasive E. coli disease (IED), including bacteraemia and sepsis, with older populations at increased risk. We aimed to estimate the theoretical coverage rate by the ExPEC4V and 9V vaccine candidates. In addition, we aimed at better understanding the diversity of E. coli isolates, including their genetic and phenotypic antimicrobial resistance (AMR), sequence types (STs), O-serotypes and the bacterial population structure.
Methods
Blood and urine culture E. coli isolates (n = 304) were collected from hospitalized patients ≥60 years (n = 238) with IED during a multicentric, observational study across three continents. All isolates were tested for antimicrobial susceptibility, O-serotyped, whole-genome sequenced and bioinformatically analysed.
Results
A large diversity of STs and of O-serotypes were identified across all centres, with O25b-ST131, O6-ST73 and O1-ST95 being the most prevalent types. A total of 45.4% and 64.7% of all isolates were found to have an O-serotype covered by the ExPEC4V and ExPEC9V vaccine candidates, respectively. The overall frequency of MDR was 37.4% and ST131 was predominant among MDR isolates. Low in-patient genetic variability was observed in cases where multiple isolates were collected from the same patient.
Conclusions
Our results highlight the predominance of MDR O25b-ST131 E. coli isolates across diverse geographic areas. These findings provide further baseline data on the theoretical coverage of novel vaccines targeting E. coli associated with IED in older adults and their associated AMR levels.
Introduction
Escherichia coli is the most common cause of Gram-negative infections in humans, possessing the ability to infect extra-intestinal sites such as the urinary tract as well as normally sterile body sites resulting in invasive E. coli disease (IED) that includes acute infections such as bacteraemia, severe pyelonephritis and sepsis. Although IED affects all age categories, the incidence increases by age with adults aged 60 years or older having an increased risk of developing the disease. Case-fatality rates range from 13% to 19% but may reach up to 60% in elderly with healthcare-associated infections.1–3 The increase in antimicrobial resistance (AMR) among E. coli strains, such as E. coli O-serotype O25b:H4-ST131 represents a major challenge for the treatment and management of E. coli infections.4 Global morbidity and hospitalization due to IED are substantial and are expected to further increase driven by ageing populations and high overall AMR prevalence.1
The O-antigen, a component of the surface polysaccharide is the target of the serum’s opsonophagocytic killing activity that elicits an immunogenic response in humans. The O-antigen provides a protective mechanism also exploited in other polysaccharide-based bacterial vaccines5,6 and has been since many years a promising target for vaccine exploration in E. coli.
A four-valent (ExPEC4V)7 and nine-valent (ExPEC9V)8 bioconjugate vaccine containing four O-antigen polysaccharides (O-serotypes O1, O2, O6 and O25) and nine O-antigen polysaccharides (O-serotypes O1, O2, O4, O6, O15, O16, O18, O25 and O75), respectively, are assessed in clinical studies towards development of a vaccine for prevention of IED.9
To estimate the O-serotype distribution of extra-intestinal pathogenic E. coli in patients with IED aged ≥60 years, a multicentre, prospective, observational study EXPECT-2 (NCT04117113) was conducted. Clinical results within this study reported by Doua et al.10 found that out of patients with IED aged ≥60 years in eight participating hospitals from seven countries, 80.4% had bacteraemic IED and half of infections was community-acquired, with the urinary tract the most common source of infection. Sepsis and septic shock were reported in 72.1% and 10.0% of patients, respectively. Kidney dysfunction was the most common complication (12.9%) and overall in-hospital mortality was 4.6%.
This study is part of the EXPECT-2 research project, complementing the clinical results reported by Doua et al.10 with a microbiological analysis of the available isolates from 238 patients with IED. The major objective was prediction of the theoretical coverage of vaccine candidates ExPEC4V and ExPEC9V based on E. coli isolates obtained from hospitalized patients aged ≥60 years with a diagnosis of IED. The O-serotype, ST, AMR profile and genomic diversity of the isolates were obtained.
Methods
Ethics
This study was conducted in accordance with the Declaration of Helsinki, Good Clinical Practices and applicable regulatory requirements. In countries where no waiver was obtained, participants or their legally acceptable representatives provided their written consent to participate in the study after having been informed about the nature/purpose of the study, and the participation and termination conditions. The protocol, Informed Consent Form, and other relevant documents were approved by an Institutional Review Board/Independent Ethics Committee (IRB/IEC) before the study was initiated.
Trial design and patient inclusion
EXPECT-2 (NCT04117113) was a prospective, observational, multicentre, hospital-based study conducted in eight participating hospitals, including two sites in Japan and one site each in the USA, Canada, France, Germany, Italy and Spain. The study took place between October 2019 and January 2021. The diagnosis of IED was determined by the microbiological confirmation of E. coli from blood, urine or an otherwise sterile body site in the presence of requisite criteria of systemic inflammatory response syndrome (SIRS), SOFA or quick SOFA (qSOFA).1
Patients were eligible for data collection if they were aged ≥60 years and hospitalized with a clinical diagnosis of IED. For countries where no waiver for informed consent had been obtained before data collection, a signed participation agreement/ICF/IAF allowing data collection and source data verification was obtained in accordance with local requirements.
All isolates from this E. coli dataset were stored in microbanks at −80°C at the local sites until shipment to the central laboratory at the University of Antwerp.
Bacterial identification
Control of viability of the stored isolates was performed by culture at the central laboratory. Identification at species level was confirmed by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany).
Antimicrobial susceptibility testing (AST)
The MIC of 24 antimicrobial agents (Table S2, available as Supplementary data at JAC Online), covering 10 different drug classes, were determined for the full dataset by microdilution method using a commercial panel (Sensititre Gram-negative GN6F AST plate, ThermoFisher Scientific). Reading of MIC values was performed with a semi-automated digital MIC viewing system (Vizion™, ThermoFisher) and results were interpreted according to the 2023 (v.13.0) EUCAST criteria.11 MDR was defined as non-susceptibility to at least one agent in three or more antimicrobial categories.12
Determination of O-serotypes
The O-serotype was determined as a combination of in silico O-genotyping and agglutination O-serotyping. O-genotyping was conducted based on WGS with identification of unique O-serotype specific sequences of wzy, wzx, wzt and wzm genes following guidelines and using reference sequences13 and were determined with the O-serotyper v.0.1 (Janssen Vaccines & Prevention, Leiden, the Netherlands).14 In case of negative O-genotyping results or in instances where the in silico result corresponded to an O-serotype belonging to one of the nine following serotypes included in the EXPEC9V vaccine candidate: O1, O2, O4, O6, O15, O16, O18, O25, O75, an O-serotyping agglutination assay (SSI Diagnostica, Hillerød, Denmark) was performed to confirm the O-genotyping result.
Whole-genome sequencing
For Illumina short-read sequencing, genomic DNA was isolated from all isolates using the Lucigen MasterPure Complete DNA and RNA Purification kit, according to the manufacturer’s protocol. Sample and library preparation was done using Nextera XT kits (Illumina Inc, San Diego, CA, USA). Libraries per isolate were pooled and adjusted to equal molar quantities. Sequencing was done using MiSeq v.2, 500 cycle, PE 2 × 250 bp (Illumina Inc., USA). Raw sequencing data were quality-assessed using FastQC, cleaned using Trimmomatic v.0.4.2 with default parameters for adapter removal and quality trimming. Contamination (<5%) and completeness (>95%) were confirmed with CheckM v.1.1.2 (taxonomy_wf species ‘Escherichia coli’).
For PacBio long-read sequencing, genomic DNA was isolated from two isolates collected from the same patient (EXP200133 and EXP200135) and selected for long-read sequencing due to major differences in AMR genes and AST, using the Qiagen® MagAttract® HMW kit (Qiagen) according to the manufacturer’s protocol. Isolated DNA was sheared using Covaris G-tubes to obtain fragments with size distributions around 8–12 kb. Barcoded SMRT bell™ libraries were prepared with the SMRTbell Template kit. Sequencing was performed using the PacBio Sequel (Pacific Biosciences, CA, USA) with 2 h pre-extension and 10 h movie time in V3 SMRTCell (1M). PacBio sequencing data were processed, demultiplexed and assembled using SMRTLink v.10.2.
Assembly, annotation and resistome analysis
Generation of the assemblies, annotations and resistomes was done using an in-house developed bacterial WGS pipeline, BacPipe v.1.2.6.15 Using this pipeline, de novo genome assemblies were made using SPAdes v.3.13.0 and genome assembly quality including number of contigs, total length and N50, was checked by QUAST v.5.0.2. Minimal average full depth coverage needed to be 50 times. Assemblies were annotated using Prokka v.1.11.1. The resistome was determined using the ResFinder v.2.1, CARD v.5.1 and ResFams v.1.2 databases with default parameters (90% identity and 60% coverage). Mutations in genes conferring resistance to fluoroquinolones (gyrA and parC) were identified with SRST2 v.0.2.0.16 SNP variants between paired isolates were detected with CLC genomics workbench v.9.5.3.
Availability of WGS data
All sequence data from this study have been deposited under BioProject ID PRJNA1021534.
MLST typing, identification of phylogroups and fimbria typing
In silico MLST based on seven conserved housekeeping genes and assignment to allelic numbers and STs was performed using the Achtman MLST locus/sequence definitions database.17In silico ClermonTyping method and its associated web-interface, the ClermonTyper v.1.4.0 was used for E. coli phylogroup determination.18,19In silico Fimbria Typing was done using FimTyper v.1.1.20
Phylogenetic analysis
Parsnp v.1.7.421 was used to create a core genome alignment of the nucleotide sequences from the unique isolate collection. SNPs within the alignment were filtered with Gubbins v.2.4.122 and used to generate a phylogenetic tree with RAxML v.8.2.12,23 using the GTRGAMMA nucleotide substitution model. To obtain statistical support for the phylogeny, 1000 bootstraps were performed. The phylogenetic tree and isolate metadata were visualized with iTOL.24
Results
IED patients and collection of E. coli isolates
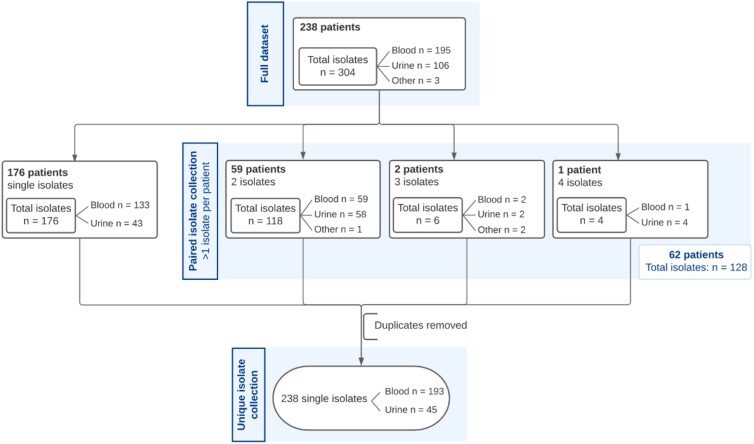
In the study, 304 E. coli isolates were included from a total of 238 unique IED patients, with 195 isolates from blood (64.1%), 106 from urine (35.0%) and three from other sterile body sites (0.9%) (Table S1). A subset of the isolates, comprising one representative E. coli isolate per IED patient, hereafter referred to as the unique isolate collection, included a total of 238 individual isolates with 193 isolated from blood and 45 from urine (Figure 1). For the dedicated analysis of paired isolates, another sub-selection was made consisting of multiple isolates per patient (n = 128 from 62 patients), hereafter called the paired isolate collection.
Figure 1.
Overview of isolate collections used for analyses. The full data collection contains 304 E. coli isolates. The unique isolate collection contains 238 isolates whereas the paired isolate collection comprises 128 isolates.
E. coli genetic diversity and dominance of O25b-ST131
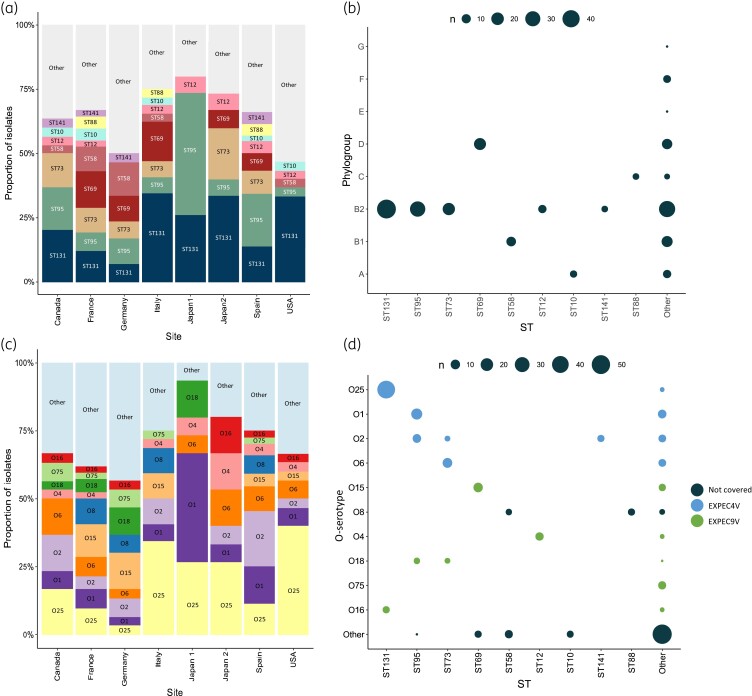
Among the unique isolate collection, genetic diversity was observed across and within study centres, resulting in the identification of 80 different STs (Figure 2a and Table S1). The nine most common STs (n = 152, 63.8%) were each found in at least five isolates and included ST131 (n = 49, 20.6%), ST95 (n = 31, 13%), ST73 (n = 19, 8%), ST69 (n = 18, 7.5%), ST58 (n = 11, 4.7%), ST12 (n = 8, 3.3%), ST10 (n = 6, 2.5%), ST88 (n = 5, 2.1%) and ST141 (n = 5, 2.1%). Seventy-one additional STs each occurred in less than five isolates from the unique isolate collection (n = 86, 36.1%). These STs were found in four isolates (one ST, n = 4, 1.7%), three isolates (four different STs, n = 12, 5%), two isolates (10 different STs, n = 20, 8.4%) or only one isolate (50 different STs, n = 50, 21%).
Figure 2.
Distribution of ST (present in ≥5 isolates) and O-serotypes (present in ≥7 isolates). STs and O-serotypes present in, respectively, five or fewer and seven or fewer isolates are summarized as ‘other’. (a) The proportion of STs across hospital sites. (b) The pairwise correlation of STs with Clermont phylogroups. The magnitude of the circles indicates the number of isolates following the legend. (c) The proportion of O-serotypes across hospital sites. D The pairwise correlation of O-serotypes and ST. The magnitude of the circles indicates the number of isolates following the legend.
Most of the 238 unique isolates belonged to phylogroup B2 (n = 147, 62%). Other identified phylogroups were phylogroup D (n = 31, 13%), B1 (n = 26, 11%), A (n = 14, 6%), C (n = 9, 4%), F (n = 7, 3%), E (n = 2, 0.8%) and G (n = 2, 0.8%). E. coli isolates from the three most found ST types (ST131, ST95, ST73) all belonged to phylogroup B2 (Figure 2b).
Forty-seven O-serotypes were detected among the unique isolates, while for nine isolates (3.8%) no O-serotype could be determined (Table S1). The six most common O-serotypes, each represented by 10 or more isolates from the collection, were O25 (n = 46, 19.3%), O1 (n = 23, 9.7%), O2 (n = 22, 9.2%), O6 (n = 17, 7.1%), O15 (n = 15, 6.3%) and O8 (n = 12, 5%) (Figure 2c). These six O-serotypes were found in more than half of all isolates (n = 135, 56.7%). Twenty-four O-serotypes were each found in one isolate (n = 24, 10%) in the collection. O-serotypes mostly correlated with specific STs (Figure 2d), with the most common correlations between O25b-ST131 (n = 44, 18%), O6-ST73 (n = 13, 5%) and O1-ST95 (n = 16, 7%).
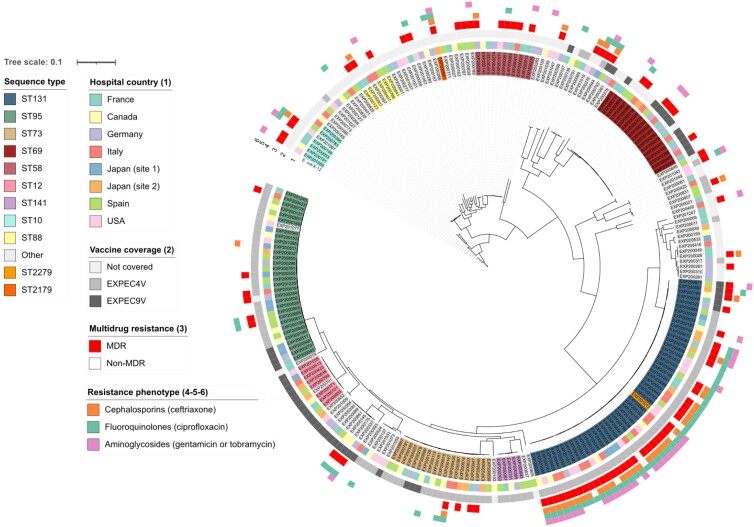
The E. coli population is genetically diverse, with the different STs presenting separate branches across the overall phylogeny (Figure 3). The dominant STs were found to be globally dispersed and there is no country-specific signature of particular STs.
Figure 3.
The population structure of extra-intestinal pathogenic E. coli is represented by a midpoint-rooted maximum-likelihood phylogenetic tree of the unique isolate collection (238 isolates), based on 1000 bootstraps. Core genomes of the isolates in this study and the reference genome E. coli K-12 were aligned. The tree is based on 139 932 SNPs. Coloured ranges represent predominant STs found in more than five isolates. Rings 1, 2 and 3 denote the hospital site, vaccine coverage by ExPEC4V and ExPEC9V, and multidrug resistance (MDR). Rings 4, 5 and 6 show resistance to cephalosporins, fluoroquinolones and aminoglycosides. The scale bar indicates the number of SNPs per site in the core genome alignment. The tree was visualized using iTOL.
Prevalence of antimicrobial resistance
Across the unique isolate collection, the highest resistance rates were reported for trimethoprim/sulfonamides (cotrimoxazole) and tetracyclines (31%, each). Substantial rates of resistance were also observed for fluoroquinolones (27.7%), third-generation cephalosporins (17.6%) and aminoglycosides (16.0%). No isolates were resistant to colistin or to carbapenems. At a geographic level, the highest proportion of resistance to third-generation cephalosporins, fluoroquinolones and aminoglycosides was observed in isolates from North America and Japan (Table 1). Eighty-nine isolates were characterized as MDR (37.4%), which were found at all eight participating sites (Table S2).
Table 1.
Prevalence of antibiotic resistance following EUCAST breakpoints
| Class of antibiotic | Europe (n = 148) | North America (n = 60) | Eastern Asia (n = 30) | Unique isolate collection (n = 238) |
|---|---|---|---|---|
| Number of isolates (%) | ||||
| Susceptible | 47 (31.8) | 23 (38.3) | 15 (50) | 85 (35.7) |
| MDRa | 54 (36.5) | 23 (38.3) | 11 (36.6) | 89 (37.4) |
| Resistance to single classb | ||||
| Aminoglycosides (gentamicin/tobramycin) | 18 (12.2) | 14 (23.3) | 6 (20) | 38 (16.0) |
| β-lactam/β-lactamase inhibitor combination (piperacillin-tazobactam) | 6 (4.1) | 4 (6.7) | 2 (6.7) | 12 (5.0) |
| Extended-spectrum cephalosporin (ceftriaxone/ceftazidime) | 19 (12.8) | 16 (26.7) | 7 (23.3) | 42 (17.6) |
| Fluoroquinolones (ciprofloxacin/levofloxacin) | 36 (24.3) | 20 (33.3) | 10 (33.3) | 66 (27.7) |
| Folate synthesis inhibitor (trimethoprim/sulfamethoxazole) | 52 (35.1) | 15 (25.0) | 7 (23.3) | 74 (31.1) |
| Tetracyclines (tetracycline/minocycline) | 52 (35.1) | 16 (26.7) | 6 (20.0) | 74 (31.1) |
| Resistance to last resort antibiotics | ||||
| Colistin | 0 | 0 | 0 | 0 |
| Ertapenem | 0 | 0 | 0 | 0 |
| Imipenem | 0 | 0 | 0 | 0 |
| Meropenem | 0 | 0 | 0 | 0 |
| Ceftazidime/avibactam | 0 | 0 | 0 | 0 |
aMDR: multidrug resistant defined as non-susceptibility to at least one agent in three of more antimicrobial categories.
bThe degree of drug resistance was based on susceptibility to representative antibiotics in the following six classes of antimicrobial drugs: aminoglycosides, extended-spectrum penicillins (piperacillin-tazobactam) β-lactam/β- lactamase inhibitors, expanded-spectrum cephalosporins, folate synthesis inhibitor and tetracyclines.
Forty of the 89 MDR E. coli belong to ST131 (n = 39) or related ST2279 (n = 1, EXP201013). These isolates are also phylogenetically related and all part of the clonal complex (CC) 131 (Figure 3). One MDR isolate from a blood specimen of a patient in Japan belonged to ST2179 (EXP200661), phylogroup B1 and was phylogenetically unrelated to the other MDR isolates. It carried a blaCTX-M-65 gene (an allelic variant belonging to CTX-M group 9) as well as a large variety of other AMR genes (16 genes, nine classes of antimicrobial agents). The ST2179 lineage has been recently reported from E. coli recovered from horses in Brazil,25 retail meat in Portugal as well as pigs and pet animals in Switzerland. While CTX-M-65-producing ST2179 E. coli had already been reported to occur in humans as commensal organisms, to our knowledge it had not yet been reported in the setting of an invasive bacterial infection in humans.
With regards to ESBL genes, apart from this particular ST2179 strain, all but one E. coli isolates of ST131/ST2279 carried a blaCTX-M-15 gene in association with blaOXA-1, aac(6’)-Ib-cr, catB3 as well as three mutations in the QRDR domain of gyrA/parC (GyrA S83L, GyrA D87N and ParC S80I) that are typically known to be associated with resistance to fluoroquinolones in E. coli. One single ST131 isolate harboured blaCTX-M-14.
Association between O-serotypes, AMR and vaccine coverage
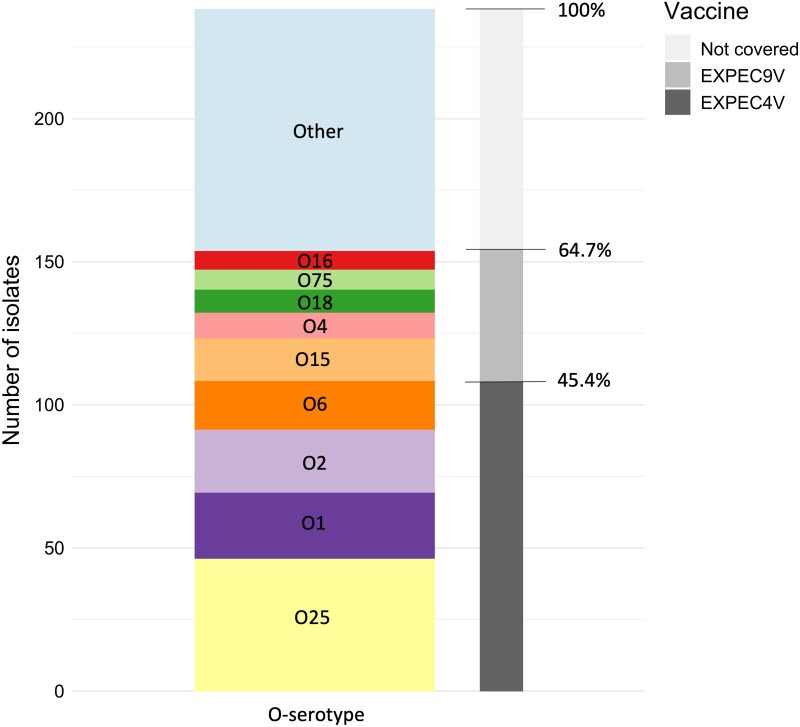
From the 238 unique isolates, 108 (45.4%) were found to carry serotypes that are included in the ExPEC4V vaccine candidate (Figure 4), whereas 154 out of 238 isolates (64.7%) contained O-antigens covered by the ExPEC9V vaccine. The most frequently found O-serotypes that were not covered by either of the vaccines were O8 (n = 12), members of the O77 group (i.e. O17/O44/O73/O77/O106, n = 6), O13 (n = 5), O9 (n = 5) and O107/O117 (n = 4).
Figure 4.
Number and ratio of isolates theoretically covered by the ExPEC4V and ExPEC9V vaccine candidate. The bar chart is coloured by the prevalence of O-serotypes in this study collection (unique isolate collection). The coverage in the ExPEC4V and ExPEC9V vaccines is coloured in shades of grey.
Out of the 89 E. coli isolates that displayed a MDR pattern, 48 (53.9%) were covered by EXPEC4V and 59 (66.3%) were covered by EXPEC9V. By far the most prevalent serotype associated with MDR E. coli was O25b always in association with ST131 that accounted for 40% of all MDR isolates (Tables S4 and S5).
Besides O25b, serotype O1 (n = 5; two in ST95, one each in ST59, ST648 and ST1177), O2 (n = 4; three in ST95, one in ST73), O15 (n = 5; four in ST69, one in ST393), O18 (n = 1; in ST1193) and O75 (n = 2; in ST1193) have all been reported previously in association with MDR E. coli isolates from human infections.4 Of note, however, 13 of the 29 (44.8%) MDR E. coli isolates carrying non-vaccine serotypes belong to well-established or emerging MDR lineages of E. coli (ST58 (n = 5), ST10/ST744 (n = 2), ST38/ST963 (n = 2), ST117, ST357, ST973, ST2179 (n = 1 each) that have been reported from animals, farm environment or food products, human intestinal carriage samples and clinical specimens from human infections.4,26,27
Analysis of paired isolates collected from blood and urine
From 62 patients, more than one isolate (n = 128/304) was collected, which we jointly called the paired isolate collection. Most of these isolates (113 isolates from 55 patients) harbour identical ST, O-serotype, AMR and AST (Table S3). For 37 isolate pairs (59.7%), SNP diversity was limited to ≤10 SNPs. Within-host diversity exceeded 10 SNPs in 25 pairs. One patient carried two O25b-ST131 isolates (EXP200133 and EXP200135) originating from a urine and blood sample, collected on the same day. In the blood isolate, six resistance genes (aac(3)-IIa, blaOXA-1, blaCTX-M-15, catB3, dfrA17, aac(6’)-Ib-cr) were detected that were not found in the urine isolate. The blood isolate also showed phenotypic resistance to nine additional antibiotics (ampicillin, ampicillin-sulbactam, aztreonam, cefazolin, cefepime, ceftazidime, ceftriaxone, gentamicin, tobramycin) as compared to the urine isolate. Long-read sequencing of both isolates from this patient revealed a plasmid carrying these resistance genes in-between transposases (Tn3 family transposase and IS6 family transposase). The full plasmid sequence (GenBank accession no. OR611935) was resolved and found to be 116 336 bp in length (Figure S1).
Discussion
Globally, the increasing burden of invasive disease due to E. coli and the continued emergence of AMR among E. coli strains is of great concern. There is a knowledge gap regarding the global distribution of E. coli ST and O-serotypes associated with IED (including sepsis and bacteraemia), with the exception of two specific studies in the UK28 and France29 describing local findings focused on bacteraemia, as well as a collection of retrospective surveillance studies describing global O-serotype epidemiology between 2011 and 201730 and genomic data from human commensal E. coli in France between 1980 and 2010.31 We present a recent, global prospective study on E. coli isolates from hospitalized IED patients combining O-typing results with phylogeny, and with a detailed comparison of AMR genotypes as determined with WGS and phenotypes with AST.
E. coli strains belonging to O25b-ST131 present a growing challenge for clinicians due to their frequent association with MDR, their increasing prevalence worldwide and ability to cause localized outbreaks of IED.30 The proportion of MDR isolates of 37% in our study of which 40% are O25, is comparable to the one reported in Oxfordshire, UK, where 44% (1434 of 3278) of the invasive E. coli isolates were found to be MDR. However, this proportion is much higher than reported by Weerdenburg et al.30 in a larger global study (with more than 3200 invasive E. coli), where approximately 10% of the isolates were classified as MDR and 60% of MDR isolates were O25. These differences might reflect regional differences, variability in MDR definition and/or antibiotic classes that were tested.
Epidemiological data about extra-intestinal E. coli in sub-Saharan Africa is still relatively scarce, but studies in Malawi,32,33 Tanzania,34 Uganda,35 South Africa36 and the Democratic Republic of Congo37 show a highly diverse E. coli population ST131 being the predominant ST.
The increasing levels of AMR among Gram-negative bacteria, including E. coli, is a global threat and is one of the main drivers for prophylactic approaches such as vaccine development. A nine-valent vaccine (EXPEC9V) for the prevention of IED is currently being tested in a pivotal vaccine efficacy trial (E.mbrace study; ClinicalTrials.gov Identifier NCT04899336). Our data support that ExPEC9V, as well as the earlier tested EXPEC4V, candidate vaccines cover a substantial number of the E. coli strains collected in this study, with a predicted vaccine coverage of approximately 45% for EXPEC4V and 65% for EXPEC9V. This finding is in line with previous findings from patients of all ages in the UK (46% and 72%; 2008–2018)28 and results from patients aged >60 years across Europe, North America, Asia-Pacific and South America (47% and 68%; 2011–2017).30 In commensal E. coli isolates from healthy individuals in France, the four O-serotypes covered by EXPEC4V represented nearly one-quarter (24%; 1980–2010) of the isolate collection.31 Previous implementation of the serotype-based pneumococcal vaccine led to an increased incidence of the serotypes that were not targeted by the vaccine.38,39 Pneumococcal vaccines induce immunity that impacts pneumococcal nasopharyngeal carriage, thereby applying a selective pressure to switch serotypes to escape the immune response. Unlike Streptococcus pneumoniae, E. coli normally inhabits the intestinal tract, representing a distinct immunological compartment. It remains unknown whether ExPEC4V and ExPEC9V apply the same selective pressure in the gut as pneumococcal vaccines do in the nasopharynx. Stool sample analysis including O-serotype prevalence and general metagenomic profiling revealed no indication of vaccine impact on E. coli prevalence, countering the notion of vaccine-induced serotype replacement.40 Close monitoring via surveillance and additional microbiome studies will be crucial to assess the risk of serotype replacement. Altogether, our findings support the further development and theoretical coverage of O-antigen based vaccines targeting E. coli. However, ongoing and future studies will need to provide evidence whether such vaccines show protective efficacy against the vaccine serotypes and assess the cost-benefit ratio of a vaccine-based prophylactic approach.
One study strength is that in a subset of patients more than one E. coli isolate was obtained from different body sites, allowing for a comparison of paired isolates from the same patient and thus assessment of the within-host diversity of the E. coli isolates both at the genomic and phenotypic levels. These data allowed us to confirm that in most of the cases the collected E. coli strains from blood and urine were genetically identical. However, in a small number of cases (four patients) different strains were observed, indicating a co-infection of multiple strains and highlighting the potential impact of within-host diversity. Alternatively, faecal contamination of the urine sample could lead to detection of an isolate that was not responsible for IED. One blood isolate harboured a plasmid with six resistance genes. Loss of this plasmid in the urine isolate compared to the blood isolate could have occurred during re-isolation and/or storage of the isolates.
Our study is part of the EXPECT-2 research project (ClinicalTrials.gov #NCT04117113) and complements the paper by Doua et al.10 in which the clinical presentation, characterization of IED by infection acquisition setting and outcome was reported in detail. Overall, the proportion of patients with community-acquired (50%) and healthcare-associated (30%) IED reflects the proportion reported in other similar studies carried out in elderly adult patients in different countries and continents.41,42
One of the limitations of our study is the potential bias of this collection by including only one enrolling site per country, except for Japan, which might not be representative of the distribution of E. coli isolates at the country level. In addition, the participating centres used their own criteria and protocols for obtaining blood culture collection, sample transport and storage of isolates. Further, a selection bias at the patient level cannot be excluded as the proportion of community- versus healthcare-acquired infection, comorbidities and severity of infection at time of enrolment could possibly have varied at the different sites. Moreover, this study was a prospective study of relatively short duration (between 2019 and 2021). This may not enable us to capture the possible emergence of new clonal lineages involved in IED or population genetic structure evolution within STs that may only appear in longitudinal surveys over longer periods (10–20 years).43–45
In conclusion, the E. coli isolates causing IED are diverse, with the most frequent being O25b-ST131, a strain with high rates of MDR. Our data showing the distribution of the phylogeny, STs, O-serotypes and AMR profiles of E. coli isolates may provide additional epidemiological and microbiological information and inform the development of multivalent conjugate vaccines that target E. coli O-antigens.
Supplementary Material
Acknowledgements
The COMBACTE-NET Consortium/EXPECT Study Group Authorship
The research leading to these results was conducted as part of the COMBACTE Consortium. For further information, please refer to www.COMBACTE.com. We thank all authors within the COMBACTE-NET Consortium/EXPECT Study Group: Gert Leten, Sofie Van Mieghem (Laboratory of Medical Microbiology of University of Antwerp, Belgium); Madison Violette, Sonal Munshi, Moussa Aitabi (Janssen); Anna Maria Azzini, Elda Righi, Nicola Duccio Salerno, Giuliana Lo Cascio, Eleonora Cremonini (University of Verona, Italy); Álvaro Pascual, Reinaldo Espíndola, Virginia Palomo (Hospital Universitario Virgen Macarena, Seville, Spain); Olivier Barraud (Limoges University, France); Sarah V. Walker (University Hospital of Cologne, Germany); Naomi Akai (Okitama Public General Hospital, Japan); Risa Kimura (Nagasaki Medical Center, Japan); Louis Lakatos, Killian De Blacam (Health Sciences North Research Institute, Canada); Joshua Thaden, Felicia Ruffin and Michael Dagher (Duke University, USA).
Contributor Information
Enya Arconada Nuin, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium.
Tuba Vilken, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium.
Basil Britto Xavier, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium; Department of Medical Microbiology and Infection Control, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
Joachim Doua, Janssen Research & Development, Infectious Diseases & Vaccines, Janssen Pharmaceutica, Beerse, Belgium.
Brian Morrow, Janssen Research & Development, Raritan, NJ, USA.
Jeroen Geurtsen, Bacterial Vaccines Discovery & Early Development, Janssen Vaccines & Prevention B.V., Leiden, The Netherlands.
Oscar Go, Janssen Research & Development, Raritan, NJ, USA.
Bart Spiessens, Janssen Research & Development, Infectious Diseases & Vaccines, Janssen Pharmaceutica, Beerse, Belgium.
Michal Sarnecki, Janssen Vaccines, Branch of Cilag GmbH International, Bern, Switzerland.
Jan Poolman, Bacterial Vaccines Discovery & Early Development, Janssen Vaccines & Prevention B.V., Leiden, The Netherlands.
Marc Bonten, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands; ECRAID, Utrecht, The Netherlands.
Miquel Ekkelenkamp, Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands.
Christine Lammens, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium.
Herman Goossens, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium.
Youri Glupczynski, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium.
Sandra Van Puyvelde, Laboratory of Medical Microbiology, Vaccine & Infectious Disease Institute, University of Antwerp, Antwerp, Belgium; Cambridge Institute of Therapeutic Immunology and Infectious Disease, University of Cambridge School of Clinical Medicine, Cambridge Biomedical Campus, Cambridge CB2 0AW, UK.
COMBACTE-NET Consortium/EXPECT Study Group:
Gert Leten, Sofie Van Mieghem, Madison Violette, Sonal Munshi, Moussa Aitabi, Anna Maria Azzini, Elda Righi, Nicola Duccio Salerno, Giuliana Lo Cascio, Eleonora Cremonini, Álvaro Pascual, Reinaldo Espíndola, Virginia Palomo, Olivier Barraud, Sarah V Walker, Naomi Akai, Risa Kimura, Louis Lakatos, Killian De Blacam, Joshua Thaden, Felicia Ruffin, and Michael Dagher
Funding
The University Medical Center Utrecht (UMCU, the Netherlands; https://www.umcutrecht.nl/en/) sponsored the study. Janssen Research & Development was the European Federation of Pharmaceutical Industries and Associations (EFPIA) partner. This research project received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115523 resources of which are composed of financial contribution from the European Union Seventh Framework Program (FP7/2007-2013), and EFPIA companies in kind contribution. The UMC Utrecht and Janssen Research & Development played a role in study design, data collection and analysis, decision to publish and preparation of the manuscript.
Transparency declarations
J.D.: Employee and stock option owner of Janssen Research & Development, a pharmaceutical company of Johnson & Johnson. B.M.: Employee and stock (option) owner of Janssen Research & Development, a pharmaceutical company of Johnson & Johnson. J.G.: Employee and stock (option) owner of Janssen Vaccines & Prevention, a pharmaceutical company of Johnson & Johnson. O.G.: Employee of Janssen Research & Development, a pharmaceutical company of Johnson & Johnson and a shareholder of Johnson & Johnson. B.S.: Employee and share owner of Johnson & Johnson. M.S.: Employee and unrestricted share owner of Janssen Vaccines, Branch of Cilag GmbH International, a Johnson & Johnson company. J.P.: Employee and stock option owner of Janssen Research & Development, a pharmaceutical company of Johnson & Johnson. M.B.: Grants paid to University Medical Center Utrecht (UMCU) from CureVac, Janssen Vaccines, Merck and Novartis; honoraria payments to MJMB from Takeda; participation on a Data Safety Monitoring Board supported by AstraZeneca, Merck, Novartis, Pfizer, Sanofi and Spherecydes. All other authors: Nothing to declare. Author(s) J.D., O.G., J.G., B.S., M.S. and J.P. belong to EFPIA (European Federation of Pharmaceutical Industries and Association) member companies in the Innovative Medicines Initiative Joint Undertaking (IMI JU), and costs related to their part in the Development were carried by the respective company as in kind contribution under the IMI JU scheme.
Supplementary data
Figure S1 and Tables S1 to S5 are available as Supplementary data at JAC Online.
References
- 1. Doua J, Geurtsen J, Rodriguez-Baño Jet al. . Epidemiology, clinical features, and antimicrobial resistance of invasive Escherichia coli disease in patients admitted in tertiary care hospitals. Open Forum Infect Dis 2023; 10: ofad026. 10.1093/ofid/ofad026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laupland KB, Gregson DB, Church DLet al. . Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clin Microbiol Infect 2008; 14: 1041–7. 10.1111/j.1469-0691.2008.02089.x [DOI] [PubMed] [Google Scholar]
- 3. Bonten M, Johnson JR, van den Biggelaar AHJet al. . Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin Infect Dis 2020; 72: 1211–9. 10.1093/cid/ciaa210 [DOI] [PubMed] [Google Scholar]
- 4. Manges AR, Geum HM, Guo Aet al. . Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev 2019; 32:e00135-18. 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cross A, Artenstein A, Que Jet al. . Safety and immunogenicity of a polyvalent Escherichia coli vaccine in human volunteers. J Infect Dis 1994; 170: 834–40. 10.1093/infdis/170.4.834 [DOI] [PubMed] [Google Scholar]
- 6. Geurtsen J, de Been M, Weerdenburg Eet al. . Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol Rev 2022; 46:fuac031. 10.1093/femsre/fuac031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janssen Research & Development L . A Study to Evaluate Safety and Immunogenicity of the ExPEC4V Clinical Trial Material After a Single Intramuscular Dose and a Second Dose 6 Months Later in Healthy Participants Aged 18 Years and Older. https://clinicaltrials.gov/ct2/show/NCT03500679
- 8. Janssen Research & Development L . A Study of Vaccination With 9-Valent Extraintestinal Pathogenic Escherichia Coli Vaccine (ExPEC9V) in the Prevention of Invasive Extraintestinal Pathogenic Escherichia Coli Disease in Adults Aged 60 Years and Older With a History of Urinary Tract Infection in the Past 2 Years. https://clinicaltrials.gov/ct2/show/NCT04899336
- 9. Fierro CA, Sarnecki M, Doua Jet al. . Safety, reactogenicity, immunogenicity, and dose selection of 10-valent extraintestinal pathogenic Escherichia coli bioconjugate vaccine (VAC52416) in adults aged 60-85 years in a randomized, multicenter, interventional, first-in-human, phase 1/2a study. Open Forum Infect Dis 2023; 10: ofad417. 10.1093/ofid/ofad417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doua J, Rodríguez-Baño J, Froget Ret al. . Clinical presentation and antimicrobial resistance of invasive Escherichia coli disease in hospitalized older adults: a prospective multinational observational study. Infection 2024; 52: 1073–85. 10.1007/s15010-023-02163-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters v.13.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.0_Breakpoint_Tables.xlsx
- 12. Magiorakos AP, Srinivasan A, Carey RBet al. . Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 13. DebRoy C, Fratamico PM, Yan Xet al. . Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS ONE 2016; 11: e0147434. 10.1371/journal.pone.0147434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iguchi A, Iyoda S, Kikuchi Tet al. . A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res 2014; 22: 101–7. 10.1093/dnares/dsu043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xavier BB, Mysara M, Bolzan Met al. . BacPipe: a rapid, user-friendly whole-genome sequencing pipeline for clinical diagnostic bacteriology. iScience 2020; 23: 100769. 10.1016/j.isci.2019.100769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inouye M, Dashnow H, Raven LAet al. . SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014; 6: 90. 10.1186/s13073-014-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3: 124. 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beghain J, Bridier-Nahmias A, Le Nagard Het al. . ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 2018; 4: e000192. 10.1099/mgen.0.000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clermont O, Dixit OVA, Vangchhia Bet al. . Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ Microbiol 2019; 21: 3107–17. 10.1111/1462-2920.14713 [DOI] [PubMed] [Google Scholar]
- 20. Roer L, Tchesnokova V, Allesøe Ret al. . Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 2017; 55: 2538–43. 10.1128/JCM.00737-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Treangen TJ, Ondov BD, Koren Set al. . The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014; 15: 524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Croucher NJ, Page AJ, Connor TRet al. . Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2014; 43: e15. 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 2021; 49: W293–W6. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leigue L, Warth JF, Melo LCet al. . MDR ST2179-CTX-M-15 Escherichia coli co-producing RmtD and AAC(6’)-Ib-cr in a horse with extraintestinal infection, Brazil. J Antimicrob Chemother 2015; 70: 1263–5. 10.1093/jac/dku520 [DOI] [PubMed] [Google Scholar]
- 26. Day MJ, Hopkins KL, Wareham DWet al. . Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 2019; 19: 1325–35. 10.1016/S1473-3099(19)30273-7 [DOI] [PubMed] [Google Scholar]
- 27. Nagy BJ, Balázs B, Benmazouz Iet al. . Comparison of extended-spectrum beta-lactamase-producing Escherichia coli isolates from rooks (Corvus frugilegus) and contemporary human-derived strains: a one health perspective. Front Microbiol 2021; 12: 785411. 10.3389/fmicb.2021.785411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lipworth S, Vihta KD, Chau KKet al. . Ten years of population-level genomic Escherichia coli and Klebsiella pneumoniae serotype surveillance informs vaccine development for invasive infections. Clin Infect Dis 2021; 73: 2276–82. 10.1093/cid/ciab006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Royer G, Clermont O, Condamine Bet al. . O-antigen targeted vaccines against Escherichia coli may be useful in reducing morbidity, mortality, and antimicrobial resistance. Clin Infect Dis 2021; 74: 364–6. 10.1093/cid/ciab458 [DOI] [PubMed] [Google Scholar]
- 30. Weerdenburg E, Davies T, Morrow Bet al. . Global distribution of O serotypes and antibiotic resistance in extraintestinal pathogenic Escherichia coli collected from the blood of patients with bacteremia across multiple surveillance studies. Clin Infect Dis 2023; 76: e1236–43. 10.1093/cid/ciac421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marin J, Clermont O, Royer Get al. . The population genomics of increased virulence and antibiotic resistance in human commensal Escherichia coli over 30 years in France. Appl Environ Microbiol 2022; 88: e0066422. 10.1128/aem.00664-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tegha G, Ciccone EJ, Krysiak Ret al. . Genomic epidemiology of Escherichia coli isolates from a tertiary referral center in Lilongwe, Malawi. Microbial Genomics 2021; 7:mgen000490. 10.1099/mgen.0.000490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Musicha P, Feasey NA, Cain AKet al. . Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. Journal of Antimicrobial Chemotherapy 2017; 72: 1602–9. 10.1093/jac/dkx058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sonda T, Kumburu H, van Zwetselaar Met al. . Whole genome sequencing reveals high clonal diversity of Escherichia coli isolated from patients in a tertiary care hospital in Moshi, Tanzania. Antimicrob Resist Infect Control 2018; 7: 72. 10.1186/s13756-018-0361-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Byarugaba DK, Erima B, Wokorach Get al. . Resistome and virulome of high-risk pandemic clones of multidrug-resistant extra-intestinal pathogenic Escherichia coli (ExPEC) isolated from tertiary healthcare settings in Uganda. PLoS ONE 2023; 18: e0294424. 10.1371/journal.pone.0294424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mbelle NM, Feldman C, Osei Sekyere Jet al. . The resistome, mobilome, virulome and phylogenomics of multidrug-resistant Escherichia coli clinical isolates from Pretoria, South Africa. Sci Rep 2019; 9: 16457. 10.1038/s41598-019-52859-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Irenge LM, Ambroise J, Bearzatto Bet al. . Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: characterization of phylogenomic changes, virulence and resistance genes. BMC Infect Dis 2019; 19: 137. 10.1186/s12879-019-3763-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanquet G, Krizova P, Dalby Tet al. . Serotype replacement after introduction of 10-valent and 13-valent pneumococcal conjugate vaccines in 10 countries, Europe. Emerg Infect Dis 2022; 28: 137–8. 10.3201/eid2801.210734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet 2011; 378: 1962–73. 10.1016/S0140-6736(10)62225-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frenck RW Jr, Ervin J, Chu Let al. . Safety and immunogenicity of a vaccine for extra-intestinal pathogenic Escherichia coli (ESTELLA): a phase 2 randomised controlled trial. Lancet Infect Dis 2019; 19: 631–40. 10.1016/S1473-3099(18)30803-X [DOI] [PubMed] [Google Scholar]
- 41. Hernandez-Pastor L, Geurtsen J, Baugh Bet al. . Clinical burden of invasive Escherichia coli disease among older adult patients treated in hospitals in the United States. BMC Infect Dis 2023; 23: 550. 10.1186/s12879-023-08479-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohmagari N, Choi WS, Tang H-Jet al. . Targeted literature review of the burden of extraintestinal pathogenic Escherichia coli among elderly patients in Asia Pacific regions. J Med Econ 2023; 26: 168–78. 10.1080/13696998.2023.2169447 [DOI] [PubMed] [Google Scholar]
- 43. Royer G, Darty MM, Clermont Oet al. . Phylogroup stability contrasts with high within sequence type complex dynamics of Escherichia coli bloodstream infection isolates over a 12-year period. Genome Med 2021; 13: 77. 10.1186/s13073-021-00892-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kallonen T, Brodrick HJ, Harris SRet al. . Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res 2017; 27: 1437–49. 10.1101/gr.216606.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodríguez I, Figueiredo AS, Sousa Met al. . A 21-year survey of Escherichia coli from bloodstream infections (BSI) in a tertiary hospital reveals how community-hospital dynamics of B2 phylogroup clones influence local BSI rates. mSphere 2021; 6: e0086821. 10.1128/msphere.00868-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.