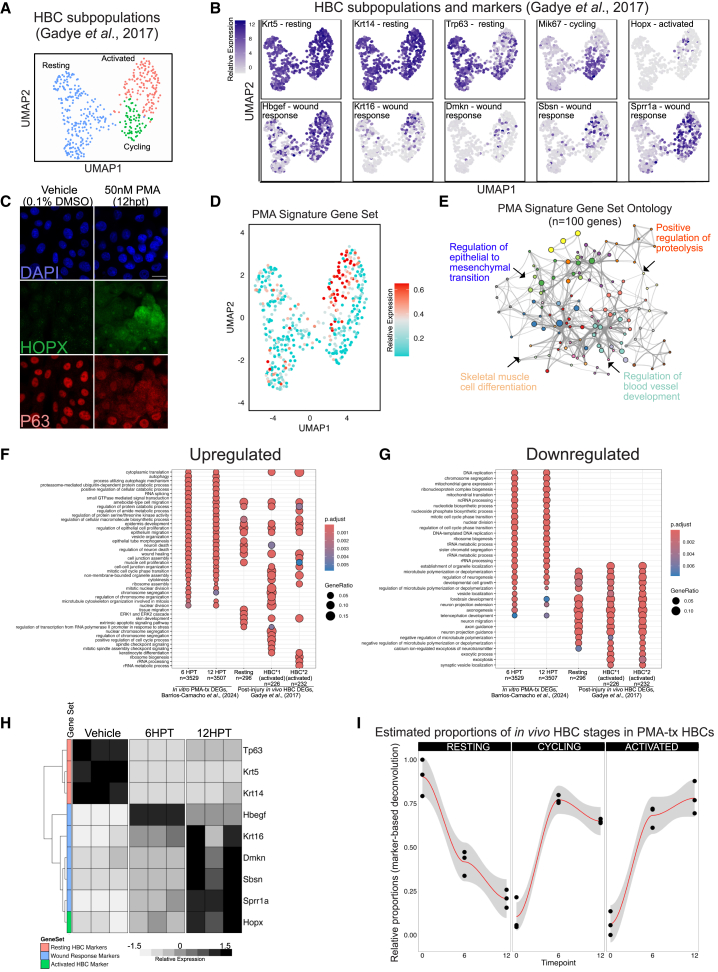
Figure 2.
PMA-treated HBCs correspond to in vivo HBCs state
(A) Clustering of HBC subpopulations found in the regenerating olfactory epithelium (Gadye et al., 2017) as described in experimental procedures.
(B) UMAP expression plot of marker genes differentiating the HBC subpopulations in vivo (from Gadye dataset): resting-HBC markers – Krt5, Krt14; dormant-HBC marker – Tp63; cycling-HBC marker – Mki67; activated HBC – Hopx; wound response markers – Hbgef, Krt16, Dmskn, Sbsn, Sprr1a.
(C) Representative immunostaining for TP63 and HOPX, an activated HBC marker previously identified. Cells were treated for 12 h with PMA before staining. Scale bar: 20 μm.
(D) Relative signature expression in HBCs. Quantity reflects the collective expression of the gene set relative to a randomized control set of the same size (see Tirosh et al., 2016.). The “PMA signature” comprises the top 100 differentially upregulated genes from the highly concordant combined 6 HPT and 12 HPT in vitro datasets (see Figure S2). Only Sprr1a is common between the PMA and in vivo (n = 100) gene sets. Cyan indicates low signature expression; red indicates high signature expression.
(E) Gene ontology network visualizing and summarizing the Biological Process terms that are enriched in the 100-gene PMA signature.
(F) clusterProfiler dotplot of GO Biological Process ontology comparing upregulated DEGs identified in the current manuscript to those identified in Gadye et al. (2017).
(G) clusterProfiler dotplot of GO Biological Process ontology comparing downregulated DEGs identified in the current manuscript to those identified in Gadye et al. (2017).
(H) Heatmap visualizing the expression of HBC markers and wound response genes detected in PMA-treated in vitro HBCs.
(I) Marker-based deconvolution of the bulk RNA-seq samples based on the single-cell gene expression patterns of resting, cycling, and activated HBCs in vivo. This approach relies on the raw single-cell data to generate markers segregating HBC states and then estimating their respective proportion in PMA-treated HBCs. The y axis defines the proportion of PMA-treated HBCs estimated to be either “resting,” “cycling,” or “activated.” For (F) and (G) FDR-adjusted p value <0.01. See NIHMS917517-supplement-5 for Gadye et al., gene lists. Please note that in vitro DEGs were converted to their mouse orthologs before enrichment analysis. Ontology categories were simplified utilizing the Wang method, with a similarity cutoff of 0.7.