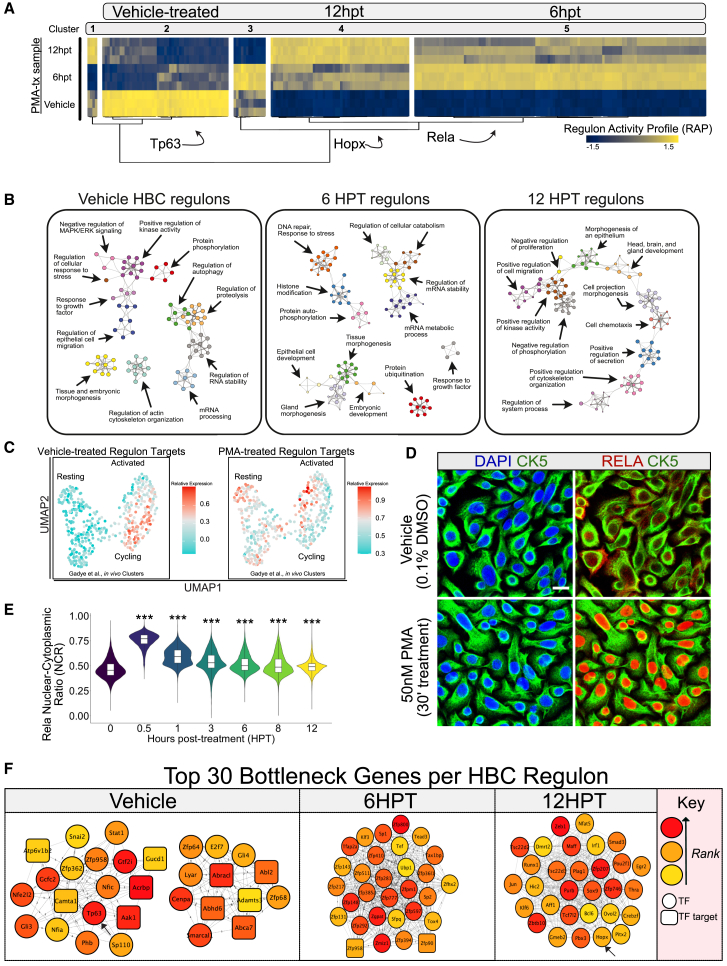
Figure 3.
Regulon analysis of PMA-treated HBCs reveals distinct networks underlying activation
(A) A heatmap representing the regulon activity profiles exported from RTN. Each vertical line represents an individual regulon (TF + target genes). Clustering is generated using Ward’s minimum variance method with Pearson correlations used as the distance used for clustering.
(B) Metascape enrichment of regulon clusters.
(C) Regulon target gene abundances from Figure 3A mapped onto the single-cell HBC data from Figure 2A for both vehicle-treated and PMA-treated transcription factors. Red indicates high expression and cyan indicates low expression.
(D) Representative immunofluorescence detailing RelA translocation 30′ after PMA treatment. Scale bar: 20 μm.
(E) An integrated violin and boxplot representing the nuclear-cytoplasmic index of RelA fluorescence over a 12-h long PMA time course (n = 3). One-way ANOVA followed by Dunnett’s Test.
(F) The top 30 bottleneck genes identified per regulon cluster. Upper left legend indicates rank of genes, as well as whether a gene is a transcription factor (circle) or a transcription factor target (square). The arrows at vehicle and at 12 HPT indicate Tp63 and Hopx, respectively.