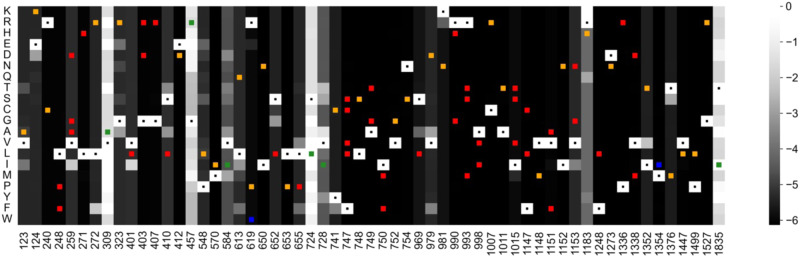
Fig. 4. Heatmap of disease-associated CACNA1D mutations with the new classifications.
The heatmap of epistatic scores shows consequences of amino acid changes to different residues at positions of reported disease-associated CACNA1D variants [11, 12]. Each column denotes one position while rows represent different amino acid residues at the corresponding position. Resulting boxes are colored according to the epistatic score of the sequence. The darkness reflects the statistical energy for the mutant to appear, i.e., the severity of the predicted impact. As an example, a dark gray box represents a rather unlikely mutant, i.e., predicted with highly negative impact. A lighter box represents a more evolutionarily favorable mutation, i.e., with less negative epistatic score. Boxes corresponding to the reference wildtype sequence are always white and noted with dots. Disease-associated CACNA1D variants are marked with squares with colors corresponding to annotations on pathogenicity. Variants were classified into pathogenic (red), likely pathogenic (orange), of uncertain significance (blue) and likely benign (green), following Table 1.