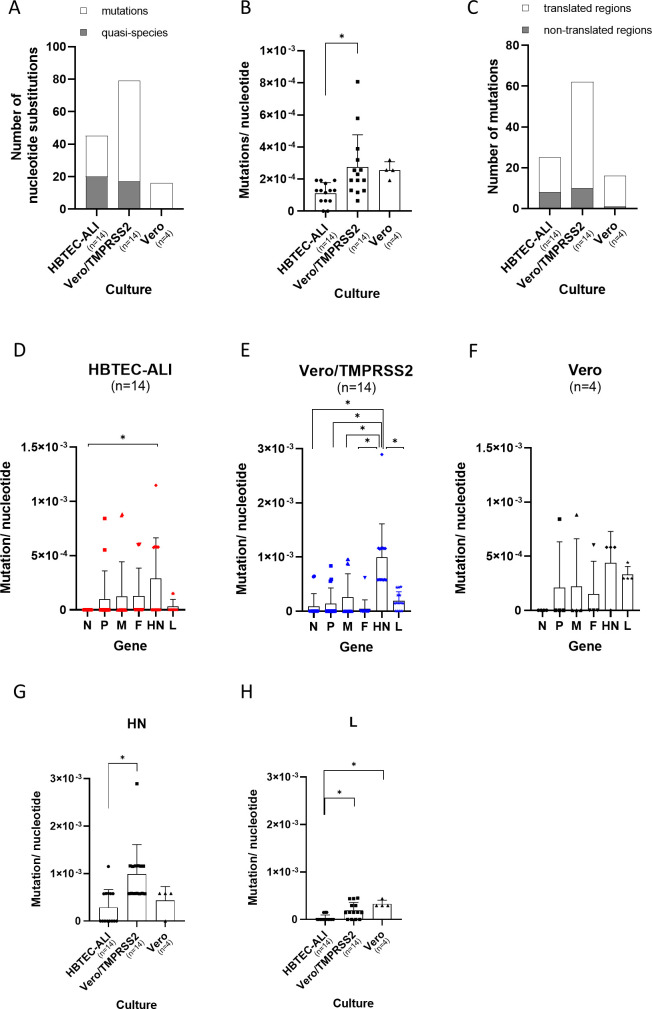
Fig 1.
Mutation rate when HPIVs were passaged in each culture. (A) Total number of nucleotide substitutions with ≥10% frequency in P5 sequences are shown. Mutations in P5 viruses were defined as those in which quasispecies were excluded from the nucleotide substitutions. The quasispecies were defined as variants that were present at ≥1% frequency in the P0 inoculum. (B) Mutation rate of each isolate after five blind passages in different cultures. (C) Regions of mutations observed in P5 sequences. (D to F) Mutation rate of each gene after five blind passages in (D) HBTEC-ALI, (E) Vero/TMPRSS2, and (F) Vero cells. (G and H) Mutation rate of (G) HN and (H) L gene in each culture. “n” indicates the number of isolates used in the study. Statistics were calculated using Tukey’s multiple comparisons test (*P < 0.05).