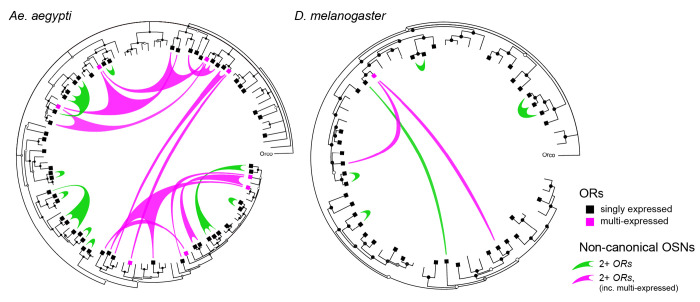
Figure 6. Patterns of OR coexpression in Ae. aegypti and D. melanogaster antennae.
Inward circle phylogenies show the evolutionary relationships among ORs found in the genomes of the dengue mosquito (left) and vinegar flies (right). Squares mark genes expressed in antennae. Green and pink ‘sails’ represent non-canonical OSN subtypes (i.e. those that express multiple receptors) and extend fingers to each receptor expressed therein, with colors distinguishing subtypes that do or do not express at least one multi-expressed gene (as in Fig. 4A). Taken together, coexpression is much less common in vinegar flies than in the dengue mosquito (fewer sails in tree on right), but shows the same signatures of coexpression by descent among singly expressed receptors (green sails tend to connect closely related ORs) and coexpression by co-option involving multi-expressed receptors (pink sails crisscross the center to connect distant ORs). Trees inferred from protein sequences using the program BAli-Phy (see Methods). Circles at tree nodes indicate bootstrap support: open >0.5, grey >0.7, black >0.9. Receptor expression data for Drosophila antennae taken from3,44,45.