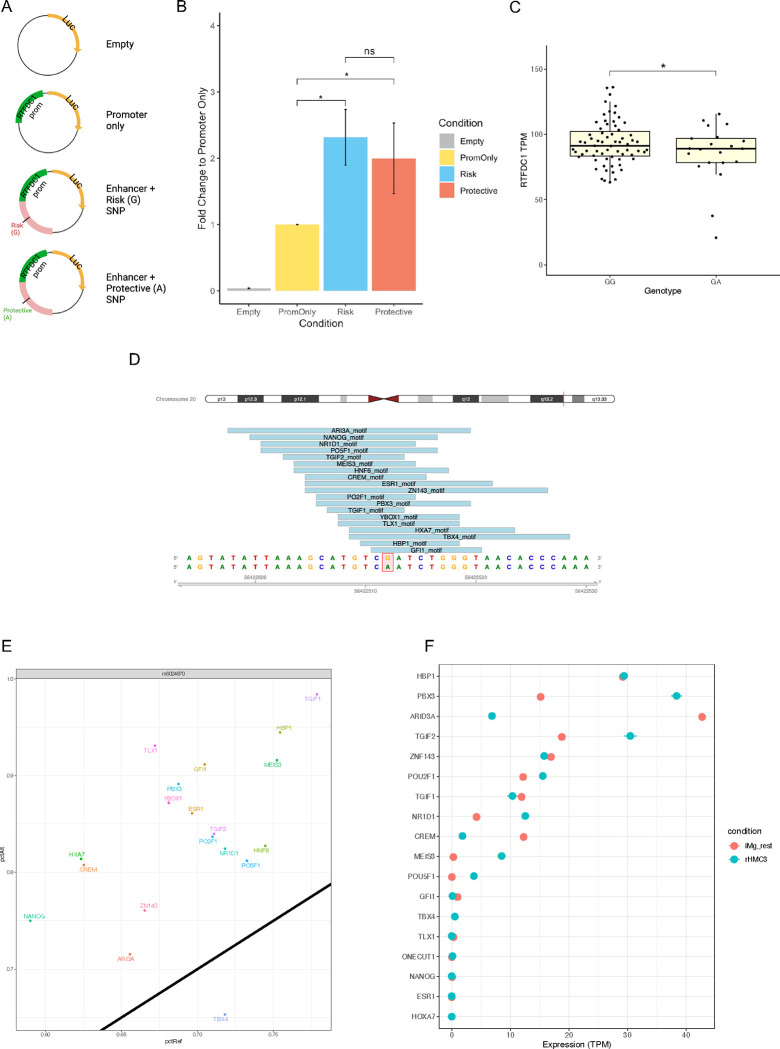
Figure 6: rs6024870 influences RTFDC1 expression in microglial cells.
(A) Schematic of plasmid design used for luciferase experiments. (B) Luciferase assay luminescence fold change of rs6024870-harboring regulatory element in HMC3 cells. N=7 biological replicates per condition. (C) Expression analysis of RTFDC1 TPMs from freshly harvested microglial cells from the FreshMicro study40. * P=0.0385 by Student’s 1-tailed T-test. (D) Transcription factor motifs overlapping rs6024870. (E) Transcription factor motif affinity analysis for rs6024870 genotype with MotifBreakR. Black diagonal line represents no change in predicted binding affinity in the presence of the protective (A) allele. Any motifs above the diagonal are predicted to have increased binding affinity in the presence of the protective allele, while motifs below the diagonal are predicted to have weakened binding affinity in the presence of the protective allele. (F) Expression of transcription factors in iMg (red) and HMC3 (blue).