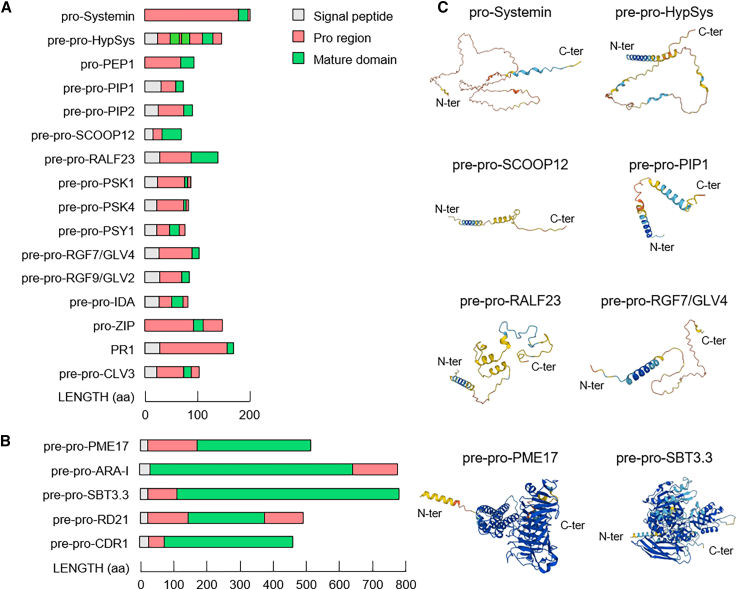
Figure 1.
Multidomain organization of protein precursors with immunomodulatory functions.
Schematic representation of the modular structures of the main (A) pro-peptides and (B) pro-enzymes discussed in the review. The lengths and positions of domains in the precursors are scaled using information obtained from UniProt (https://www.uniprot.org/). The N-terminal signal peptide (gray box), pro region (pink box), and active domain (green box) are indicated. The scales below indicate the domain lengths expressed in number of aa.
(C) AlphaFold (https://alphafold.ebi.ac.uk/) models of six representative phytocytokine precursors: pro-Systemin (UniProt accession P27058), pre-pro-HypSys (UniProt accession Q7XAD0), pre-pro-SCOOP12 (UniProt accession B3H7I1), pre-pro-PIP1 (UniProt accession Q1PE40), pre-pro-RALF23 (UniProt accession Q9LUS7), and pre-pro-RGF7/GLV4 (UniProt accession Q6NNL3); and two representative enzyme precursors: pre-pro-PME17 (UniProt accession O22149) and pre-pro-SBT3.3 (UniProt accession Q9MAP5). The AlphaFold models are shaded on the basis of the per-residue model confidence (pLDDT) score: dark blue represents the most confidently predicted regions, transitioning through light blue and yellow to orange for regions with very low confidence. N-ter, N-terminal; C-ter, C-terminal.