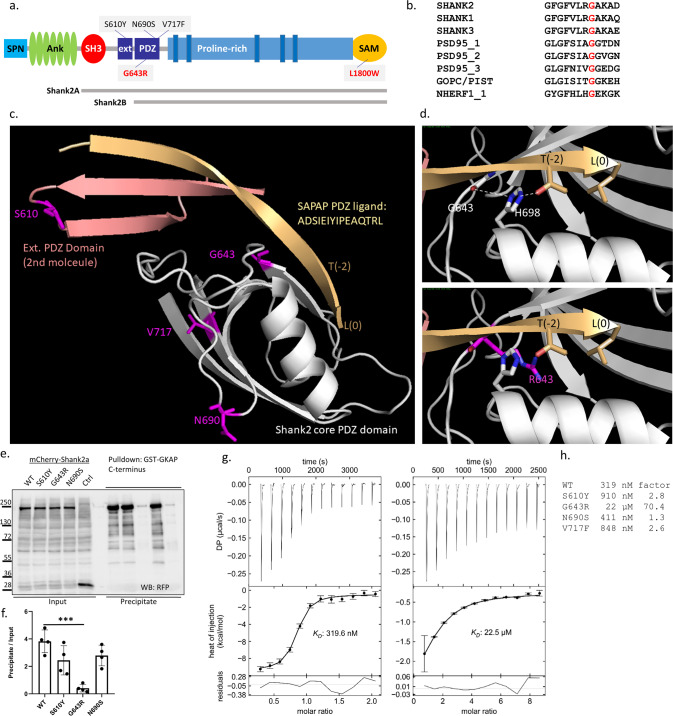
Fig. 1. Mutations affecting key domains of SHANK2.
a Domain structure of a generic Shank protein, ranging from SPN to SAM domains. The positions of variants in Shank2 PDZ and SAM domains are indicated (black for previously reported variants, red for variants reported in this study). Transcript variants Shank2a and Shank2b were used in this study and are indicated. b Alignment of the sequence around G643 in Shank2 with that of other type I PDZ domains. c Model of the extended PDZ domain of Shank2 in complex with the C-terminal PDZ ligand of GKAP/SAPAP1. Modeling is based on the structure of the Shank3/GKAP complex (5IZU) [50]. The extended binding surface is formed by two Shank2 molecules, one of which (light gray) provides the core PDZ domain, whereas the second molecule provides the extended sequence (salmon). The C-terminal GKAP sequence forms an extended β-sheet first with the extension part provided by the second molecule, and then with a β-strand of the core PDZ domain. The C-terminal leucine L(0) and the threonine at position (−2) of the PDZ ligand are indicated. S610, G643, N690 and V717 are indicated in magenta. d Upper panel. Magnification of the binding site in the core PDZ domain. G643 is located at the C-terminal end of the β-strand which is in contact with the PDZ ligand. Its carbonyl group forms an H-bond (white dashed line) to N1 of the conserved histidine H698; N2 of H698 makes another H-bond to the side chain oxygen of T(−2). Lower panel. Model of the G643R mutant protein. The introduction of the bulky arginine at position 643 (magenta) shortens the β-strand, and slightly displaces it upwards. In addition, R643 clashes with the side chain of H698 and pushes it to the left. This displacement is predicted to disrupt the interaction of H698 with T(−2) of the PDZ ligand. e mCherry-tagged Shank2a variants were expressed in 293T cells. Cell lysates were subjected to pulldown with a GST fusion protein of GKAP C-terminus. Input and precipitate samples were analyzed by Western blotting using anti-RFP antibodies. f Quantification of precipitation; ***, significantly different from WT, p < 0.001; data from four independent experiments; ANOVA, followed by Dunnett’s multiple comparison test. g Isothermal titration calorimetry of purified His6–SUMO tagged fusion protein containing SH3 to PDZ domains of Shank2 WT (left) and G643R mutant (right) vs. the synthetic peptide ADSIEIYIPEAQTRL which corresponds to the C-terminus of GKAP/SAPAP proteins. h Comparison of binding affinities for all PDZ domain mutants. The factor on the right is calculated as the ratio of Kd values for mutant divided by WT. ITC curves for the mutants S610Y, N690S and V717F are shown in the Supplementary Fig. S2.