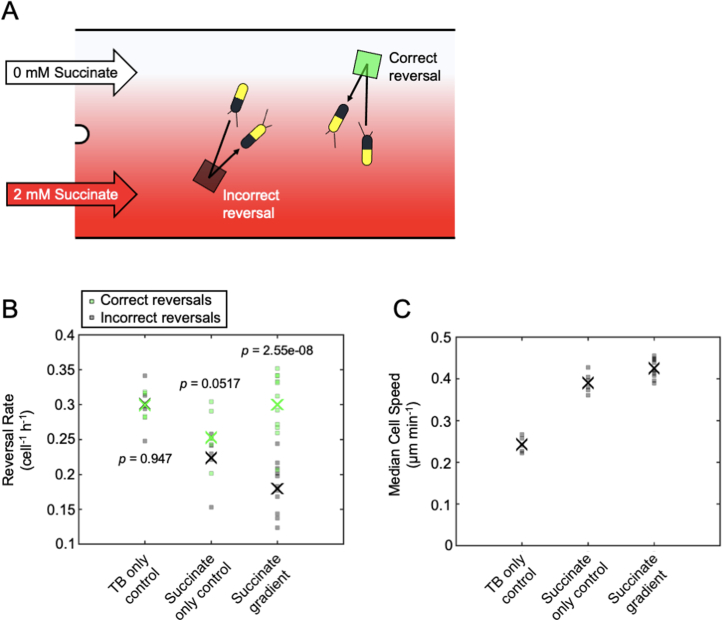
Extended Data Fig. 1. Surface-attached P. aeruginosa cells climb spatial succinate gradients by actively changing the rate at which they reverse direction.
(a) A dual-inlet BioFlux microfluidic device was used to expose cells to a spatial gradient of succinate (CMIN = 0 mM, CMAX = 2 mM) with a characteristic length-scale of 100 µm, and an automated algorithm was used to detect when cells reversed their movement direction33. Reversals were classified as either ‘correct’ or ‘incorrect’. Correct reversals (green square) occur in cells that were initially moving away from the source of succinate, whilst incorrect reversals (black square) occur in cells that were initially moving towards the source of succinate. (b) The rate of correct reversals is significantly greater than that of incorrect reversals, which drives chemotaxis towards succinate. Crosses (‘X’) mark the mean of five separate TB-only control experiments, six separate succinate-only control experiments, or eleven separate succinate gradient experiments. In TB-only experiments, TB is passed through both inlets at the same time, whilst in succinate-only experiments, media containing succinate is passed through both inlets at the same time. Controls were processed in the same way as for succinate gradients, but since no gradient was actually present, the ‘correct’ and ‘incorrect’ rates shown are arbitrary. p-values were obtained from two-sided paired t-tests of the null hypothesis that the measured incorrect and correct reversal rates come from the same distribution. (c) Cell speed is significantly higher in the presence of succinate gradients compared to both control experiments and is significantly higher in succinate-only controls compared to TB-only controls (one-way ANOVA, p = 2.22 × 10−11). The analysis in panels B and C only includes trajectory time-points that occurred before 5 h, after which the cell density becomes too high to reliably track cells in these experiments. The succinate gradient datasets additionally only include trajectory time-points above a minimum gradient strength of 0.0006 mM µm−1 to ensure that cells within regions of the device where there is a very small spatial gradient were not included in our analysis. Source data provided as a Source Data file.