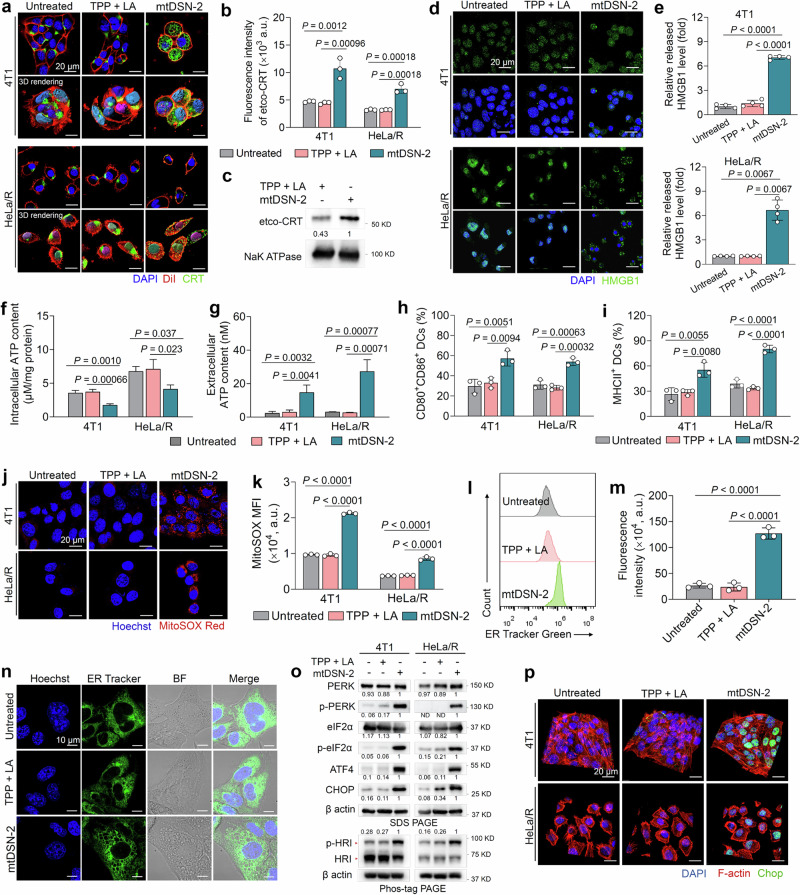
Fig. 3. mtDSN-2 incites ICD cascade related to cellular stress.
a Representative confocal microscopy images showing externalized CRT on the cell membrane after mtDSN-2 exposure. Cell membrane: DiI (red), CRT: antibody against CRT (green), nuclei: DAPI (blue). b The CRT exposure to cell surface was examined by immunofluorescence cytometry among viable cells (propidium iodine-negative). c Immunoblotting detection of CRT in the plasma membrane protein fraction. d Confocal microscopy images showed HMGB1 release from the nuclei to the cytoplasm after mtDSN-2 treatment. HMGB1: antibody against HMGB1 (green), nuclei: DAPI (blue). e Extracellular HMGB1 release determined by ELISA. Changes in intracellular (f) and extracellular (g) ATP content following different treatments. h, i Flow cytometry analysis of BMDC maturation after treatments (gated on CD11c+ dendritic cells). Confocal microscopy (j) and flow cytometry (k) analysis of mitochondrial ROS production in tumor cells by MitoSox staining. Mitochondrial Superoxide: MitoSox Red (red), nuclei: Hoechst 33342 (blue). l, m Flow cytometry analysis showing ER Tracker Green in 4T1 cells. n ER swelling-related cytoplasmic vacuolation assessed by confocal microscopy. o Western blot analysis of ER-stress related protein in cells treated with mtDSN-2 (5 µM for 36 h). In the phos-tag PAGE assay, a phos-tag gel was used to resolve the protein phosphorylation, which can be assessed by the mobility shift. Band intensities were quantified from the 8-bit digital images by densitometry in ImageJ and are shown normalized to the mtDSN-2 lane for each target. ND, not detected. p Confocal microscopy images showing CHOP expression in tumor cells after different treatments. CHOP: antibody against CHOP (green), F-actin: Rhodamine Phalloidin (red), nuclei: DAPI (blue). a.u., arbitrary units. The data are presented as the mean ± s.d. The representative images of panels (a, d, j, and p) were repeated in two biologically independent cell lines. n = 3 biologically independent experiments (b, f–i, and k–n). n = 4 biologically independent samples (e). The similar results were repeated in two biologically independent experiments (c, o). Statistical analysis by one-way ANOVA with Turkey’s multiple comparisons test. Source data are provided as a Source data file.