Abstract
On the basis of similarities between known xylanase sequences of the F family, three invariant acidic residues of xylanase A from Streptomyces lividans were investigated. Site-directed-mutagenesis experiments were carried out in Escherichia coli after engineering the xylanase A gene to allow its expression. Replacement of Glu-128 or Glu-236 by their isosteric form (Gln) completely abolished enzyme activity with xylan and p-nitrophenyl beta-D-cellobioside, indicating that the two substrates are hydrolysed at the same site. These two amino acids probably represent the catalytic residues. Immunological studies, which showed that the two mutants retained the same epitopes, indicate that the lack of activity is the result of the mutation rather than misfolding of the protein. Mutation D124E did not affect the kinetic parameters with xylan as substrate, but D124N reduced the Km 16-fold and the Vmax. 14-fold when compared with the wild-type enzyme. The mutations had a more pronounced effect with p-nitrophenyl beta-D-cellobioside as the substrate. Mutation D124E increased the Km and decreased the Vmax. 5-fold each, while D124N reduced the Km 4.5-fold and the Vmax. 75-fold. The mutations had no effect on the cleavage mode of xylopentaose.
Full text
PDF

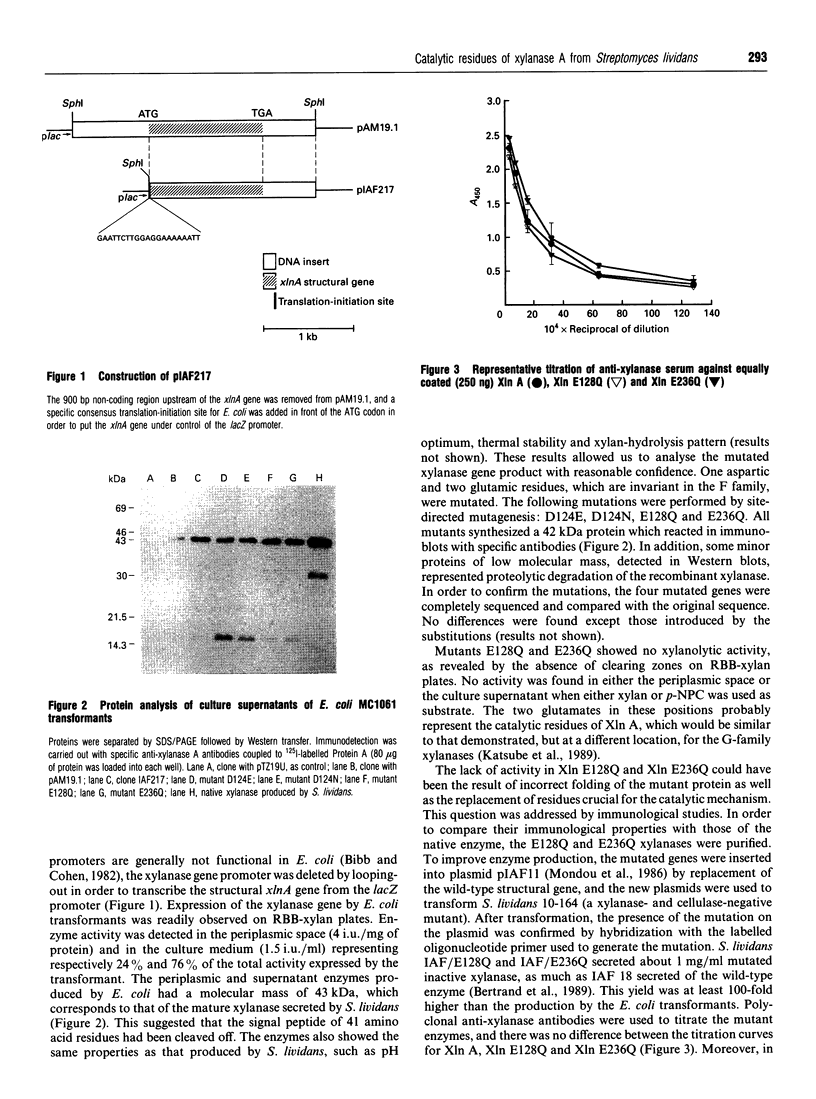

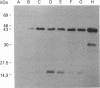
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F., Prody C., Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984 Dec;160(3):1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-beta-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Biely P., Kluepfel D., Morosoli R., Shareck F. Mode of action of three endo-beta-1,4-xylanases of Streptomyces lividans. Biochim Biophys Acta. 1993 Mar 26;1162(3):246–254. doi: 10.1016/0167-4838(93)90288-3. [DOI] [PubMed] [Google Scholar]
- Biely P., Krátký Z., Vrsanská M. Substrate-binding site of endo-1,4-beta-xylanase of the yeast Cryptococcus albidus. Eur J Biochem. 1981 Oct;119(3):559–564. doi: 10.1111/j.1432-1033.1981.tb05644.x. [DOI] [PubMed] [Google Scholar]
- Biely P., Mislovicová D., Toman R. Soluble chromogenic substrates for the assay of endo-1,4-beta-xylanases and endo-1,4-beta-glucanases. Anal Biochem. 1985 Jan;144(1):142–146. doi: 10.1016/0003-2697(85)90095-8. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Gebler J., Gilkes N. R., Claeyssens M., Wilson D. B., Béguin P., Wakarchuk W. W., Kilburn D. G., Miller R. C., Jr, Warren R. A., Withers S. G. Stereoselective hydrolysis catalyzed by related beta-1,4-glucanases and beta-1,4-xylanases. J Biol Chem. 1992 Jun 25;267(18):12559–12561. [PubMed] [Google Scholar]
- Haga K., Kitaoka M., Kashiwagi Y., Sasaki T., Taniguchi H. Purification and properties of a xylanase from Cellvibrio gilvus that hydrolyzes p-nitrophenyl cellooligosaccharides. Agric Biol Chem. 1991 Aug;55(8):1959–1967. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc Natl Acad Sci U S A. 1989 Jan;86(1):133–137. doi: 10.1073/pnas.86.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J., Grütter M. G., Anderson W. F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981 Apr 25;147(4):545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Mondou F., Shareck F., Morosoli R., Kluepfel D. Cloning of the xylanase gene of Streptomyces lividans. Gene. 1986;49(3):323–329. doi: 10.1016/0378-1119(86)90368-9. [DOI] [PubMed] [Google Scholar]
- Morosoli R., Bertrand J. L., Mondou F., Shareck F., Kluepfel D. Purification and properties of a xylanase from Streptomyces lividans. Biochem J. 1986 Nov 1;239(3):587–592. doi: 10.1042/bj2390587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podkovyrov S. M., Burdette D., Zeikus J. G. Analysis of the catalytic center of cyclomaltodextrinase from Thermoanaerobacter ethanolicus 39E. FEBS Lett. 1993 Feb 15;317(3):259–262. doi: 10.1016/0014-5793(93)81288-b. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareck F., Roy C., Yaguchi M., Morosoli R., Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991 Oct 30;107(1):75–82. doi: 10.1016/0378-1119(91)90299-q. [DOI] [PubMed] [Google Scholar]
- Sierks M. R., Ford C., Reilly P. J., Svensson B. Catalytic mechanism of fungal glucoamylase as defined by mutagenesis of Asp176, Glu179 and Glu180 in the enzyme from Aspergillus awamori. Protein Eng. 1990 Jan;3(3):193–198. doi: 10.1093/protein/3.3.193. [DOI] [PubMed] [Google Scholar]
- Takase K. Effect of mutation of an amino acid residue near the catalytic site on the activity of Bacillus stearothermophilus alpha-amylase. Eur J Biochem. 1993 Feb 1;211(3):899–902. doi: 10.1111/j.1432-1033.1993.tb17623.x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D., Withers S. G., Gilkes N. R., Kilburn D. G., Warren R. A., Aebersold R. Glutamic acid 274 is the nucleophile in the active site of a "retaining" exoglucanase from Cellulomonas fimi. J Biol Chem. 1991 Aug 25;266(24):15621–15625. [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]