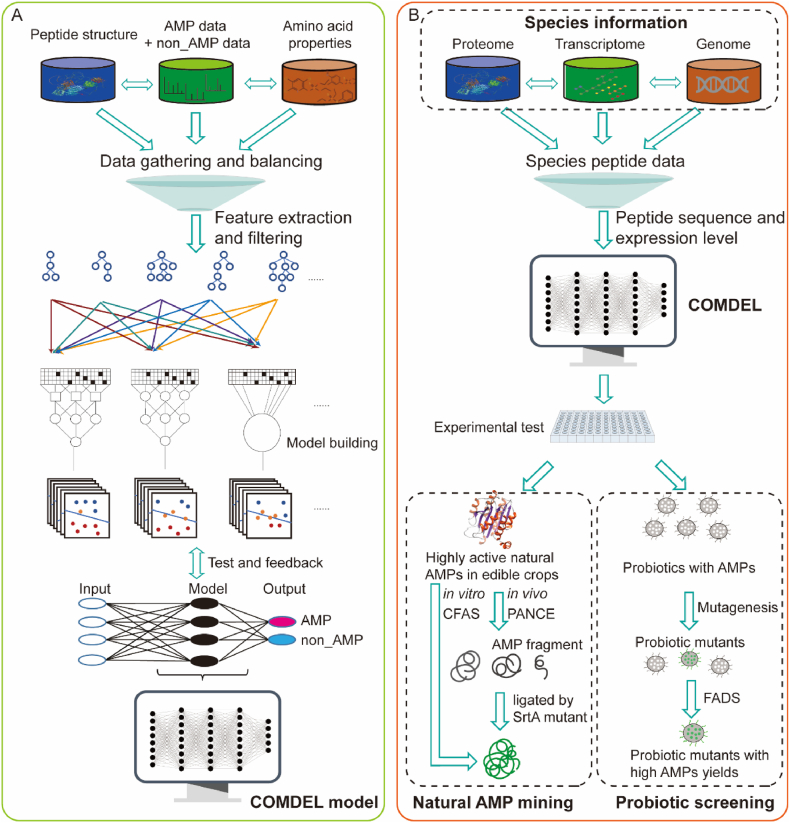
Figure 1.
The schemes of COMDEL establishment and applications. (A) Schematic depiction of COMDEL development. The AMP and non_AMP Data were collected, followed by which the features of amino acids properties and peptide structure were extracted and filtered. Through the NNA picking and feature weighting, the model was trained and tested with feedback. This process resulted in the final version of COMDEL, characterized by high accuracy and precision. (B) Application of COMDEL in mining AMPs and probiotics. For mining AMPs and probiotics, omics data encompassing genomes, transcriptomes, and proteomes were compiled. These data were processed to deduce peptide expression levels, which were then evaluated by the COMDEL model to identify potential AMP candidates for subsequent biosynthesis and validation. In the context of natural AMP mining, COMDEL was employed to pinpoint AMP candidates with high expression in edible crops. These candidates were synthesized in vivo using an SrtA mutant evolved through the PANCE technology, and in vitro via a CFAS system. In the realm of probiotic screening, COMDEL was utilized to filter edible probiotics with elevated AMP expression. Subsequently, the probiotic mutants with increased AMP yields were obtained by using the FADS technology.