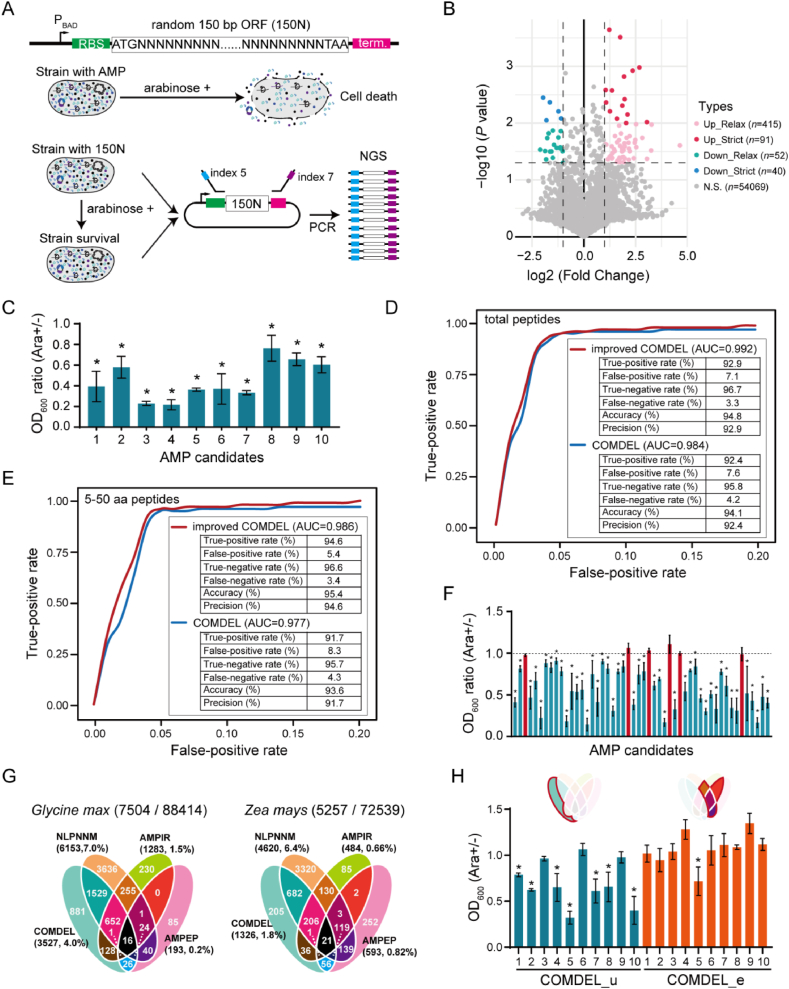
Figure 4.
Optimization and application of the COMDEL model in AMP mining. (A) Schematic diagram of the high-throughput AMP screening method. A vector pool that contains 150N DNA sequences were controlled by the arabinose operon. The presence of bacteriostatic activity in encoded peptides results in growth inhibition of the strain upon the addition of l-arabinose. The 150N DNA is then amplified using universal primers and sequenced on the Illumina NovaSeq 6000 platform. (B) The volcano plot displays the difference in peptide expression under the conditions with and without l-arabinose by analysing sequencing data. The labels “up-relax” and “up-strict” refer to peptides with an expression increase (fold change >2) under relax (P value < 0.05) and strict (P value < 0.01) conditions, while “down-relax” and “down-strict” refer to peptides with reduced expression (fold change <0.5) under relax (P value < 0.05) and strict (P value < 0.01) conditions. (C) The effect of 10 AMP candidates screened by high-throughput method on the growth of E. coli DH5α. (D and E) The AUROC curve comparison of the COMDEL models before and after optimized in total peptides (D) and the peptides shorter than 50 amino acids (E) using the unified test dataset. (F) The effect of 50 AMP candidates predicted by COMDEL from the two edible crops on the growth of E. coli DH5α. (G) The venn diagram displays the AMP candidates predicted by the optimized COMDEL model and other methods in two edible crops. A total of 7504 and 5257 AMP candidates were identified by these four models in Glycine max and Zea mays, respectively, from datasets comprising 88,414 and 72,593 peptides. The percentages represent the proportion of AMP candidates predicted by each model to the total number of peptides. (H) The effect of the AMP candidates predicted by COMDEL and other methods on the growth of E. coli DH5α. COMDEL_u represents the AMP candidates uniquely identified by the COMDEL model, while COMDEL_e represents the AMP candidates identified by at least two models as opposed to the COMDEL model. When comparing the OD600 value of the condition with and without l-arabinose, an asterisk (∗) denotes that the data are significantly different.