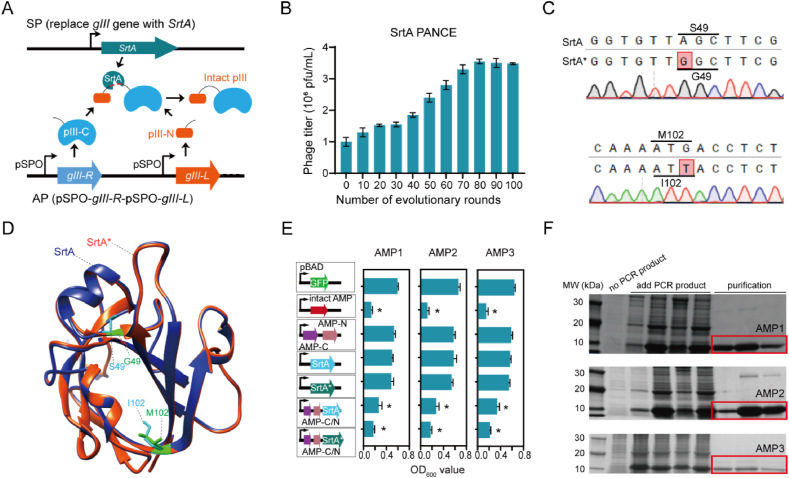
Figure 5.
Establishment of AMP biosynthesis approaches. (A) A biosensor of SrtA activity linked to peptide ligation couples with the abundance of M13 essential protein pIII. In the selection phage (SP), the gIII gene is replaced with the SrtA gene. In the accessory plasmid (AP), the gIII gene is segmented into two portions according to a recent publication52, requiring ligation by SrtA to form the full-length pIII protein. (B) Detection of the M13 phage titer change during the progression of evolutionary rounds in SrtA PANCE. (C) Sanger sequencing to detect the SrtA mutations in PANCE. (D) The structures comparison of SrtA and its mutants (SrtA∗) predicted by AlphaFold2. Alterations in the side chains of mutated amino acids are delineated and presented using ChimeraX. (E) The peptide ligation efficacy of SrtA to AMP is validated by the effect on strain growth. When comparing to the OD600 value of the GFP vector, an asterisk (∗) denotes that the data under corresponding conditions are significantly different. (F) SDS-PAGE to exhibit three AMPs identified by COMDEL from the two edible crops generated by the cell-free synthesis system, and then purified by gel and ion exchange chromatography.