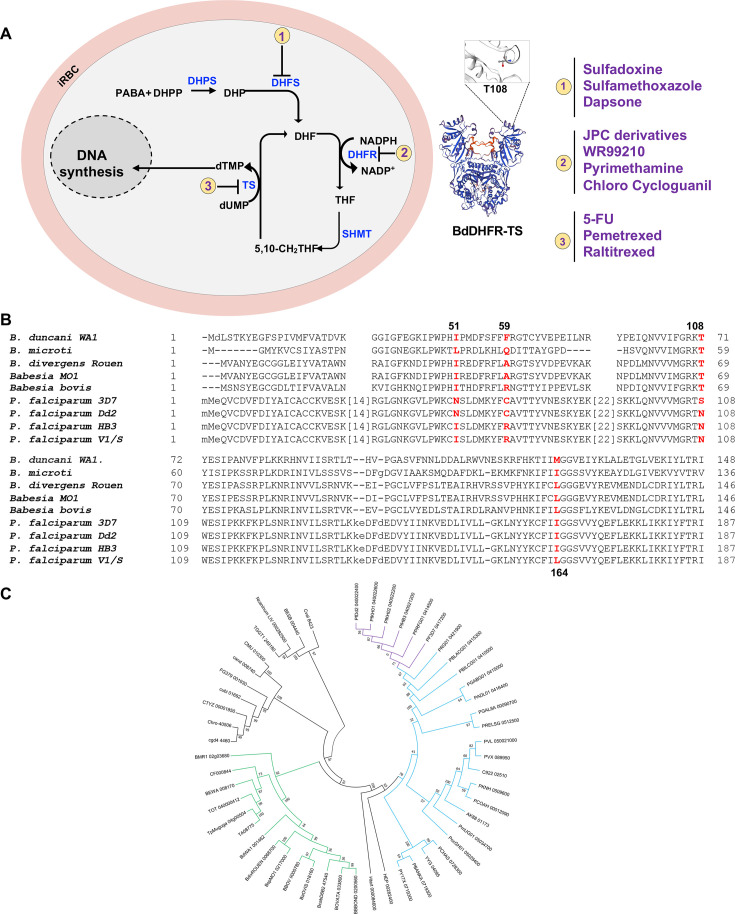
Fig 1.
Conservation of the folate metabolic pathway among Babesia and Plasmodium parasites. (A) A schematic representation of folate metabolism in Babesia and Plasmodium parasites. The main enzymes (blue color) targeted by antifolates (violet color) are highlighted. Abbreviations (Abbs.): iRBC = infected red blood cell, DHP = dihydropteroate, DHPP = dihydropteroate pyrophosphate, DHPS = dihydropteroate synthase, DHF = dihydrofolate, DHFR = dihydrofolate reductase, PABA = p-aminobenzoic acid, SHMT = Serine hydroxy methyltransferase, THF = tetrahydrofolate, and TS = thymidylate synthase. (B) Alignment of a region of DHFR-TS enzymes from various Babesia species as well as pyrimethamine-sensitive (3D7) and -resistant (Dd2, HB3, and V1/S) P. falciparum strains. Residues known to be associated with susceptibility or tolerance to antifolates are marked in red. (C) Phylogenetic analysis of DHFR-TS enzymes from piroplasmids (tick-transmitted intraerythrocytic parasites) and various Plasmodium species. The tree was built using Phylogeny.fr pipeline combining Mafft, BMGE, and FastTree software applications. An advanced option was used to generate 1,000 bootstraps providing branch support. Clade supporting Piroplasmida sequences is shown in green; Plasmodium sequences are in blue, with branches of P. falciparum sequences represented in violet.