Figure 1:
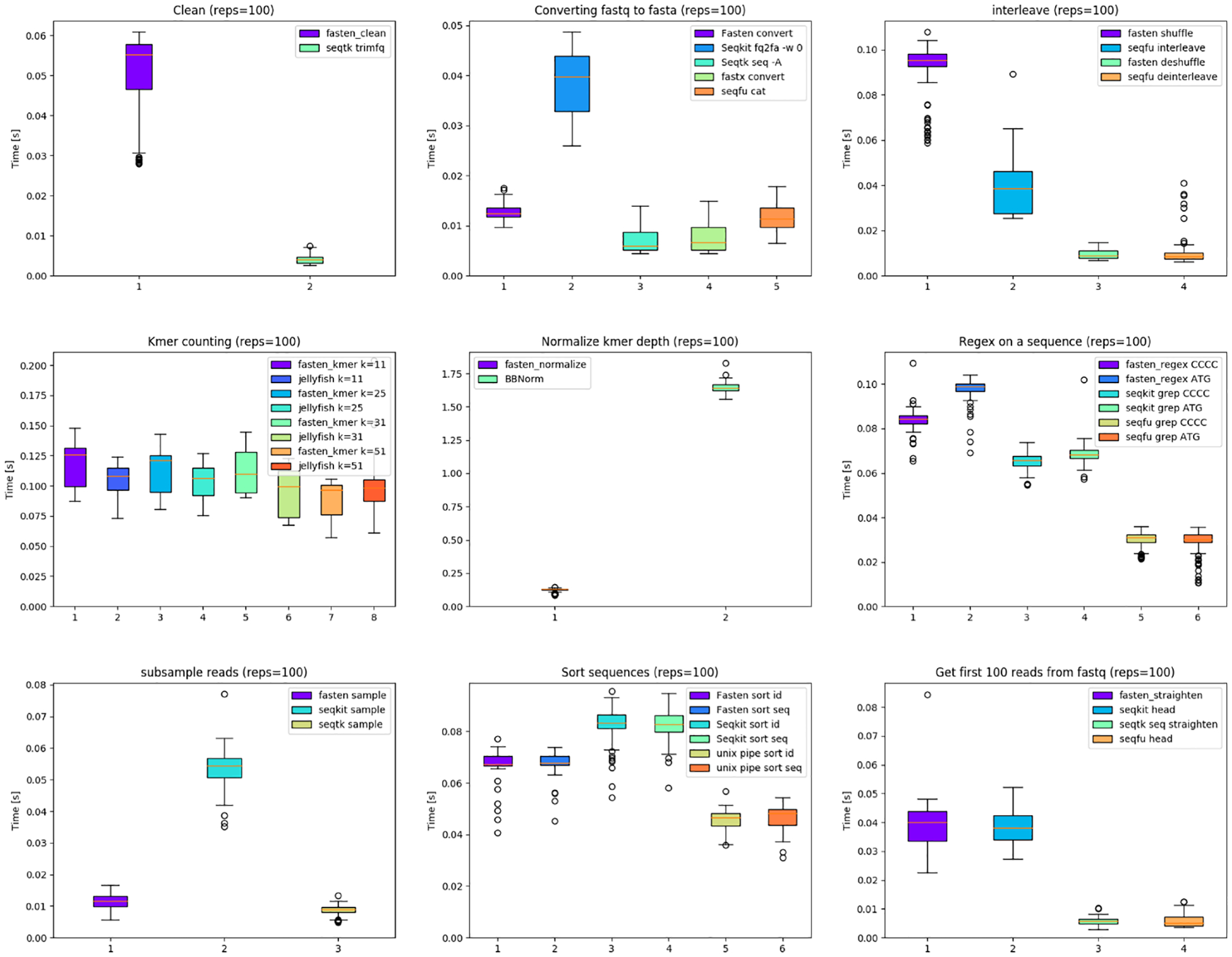
Benchmarks comparing fasten with other analogous tools. From left to right, then to bottom: Trimming with a minimum quality score; converting fastq to fasta; interleaving R1 and R2 reads; kmer counting; normalizing read depth using kmer coverage; searching for a sequence in a fastq file; downsampling reads; sorting fastq entries by either sequence or ID; and converting nonstandard fastq files to a format whose entries are four lines each, and selecting the first 100.
