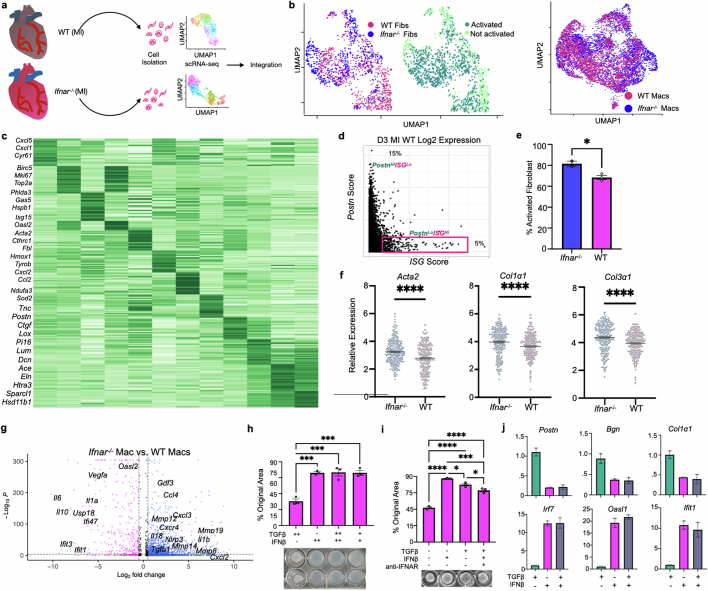
Extended Data Fig. 9. Signalling through type I IFN receptor antagonizes fibroblast activation and function.
(a) Whole cell isolation from WT and IFNAR-deficient hearts processed for scRNA-seq. Single cell datasets were integrated and log normalized (n = 4 WT, 2 Ifnar−/− mice). (b) UMAP of clustered fibroblasts and macrophages from WT and Ifnar−/− D4 infarcted hearts. Integrated fibroblasts were split by activated vs. nonactivated fibroblasts based on differential expression of Postn gene score. (c) Heatmap of fibroblast subcluster marker genes. (d) Feature scatterplot from D4 MI hearts showing an inverse scatter profile between PostnHI and ISG scores. Approximately 5% of fibroblasts isolated from WT infarcted hearts are IFNICs and express low counts of Postn (n = 4 WT mice). (e) Percentage of activated fibroblast composition between Ifnar−/− and WT mouse hearts and (f) log normalized expression of gene markers in activated fibroblast populations (n = 784 WT and 802 Ifnar−/− fibroblasts). (g) Volcano plot of differentially expressed genes between WT and Ifnar−/− macrophages corrected (n = 3628 WT macs, 3885 Ifnar−/− macs). (h) Collagen Gel contraction assays performed with L929 fibroblasts treated with 10 ng/mL TGFβ, 10 ng/mL IFNβ, or combination treatment (n = 3 replicates per condition). (i) Treatment with anti-IFNAR-ab abrogated the IFNβ-mediated blockade of the fibroblast contractile phenotype (n = 3 gels per condition). (j) Top: L929 fibroblast gene expression of matricellular proteins after treatment with TGFβ, IFNβ, or combination treatment, and corresponding inverse expression of ISG’s in the bottom panel (n = 2 replicates per condition). Data was analysed using two-tailed unpaired t or Mann-Whitney test (e, f), Wilcoxon rank sum test with Benjamini-Hochberg FDR correction (b, c, g), and two-tailed one-way ANOVA with Bonferroni’s post-hoc analysis (h, i, j). Data are mean ± s.e.m., *P > 0.05, *** P > 0.0005 ****P > 0.00005.