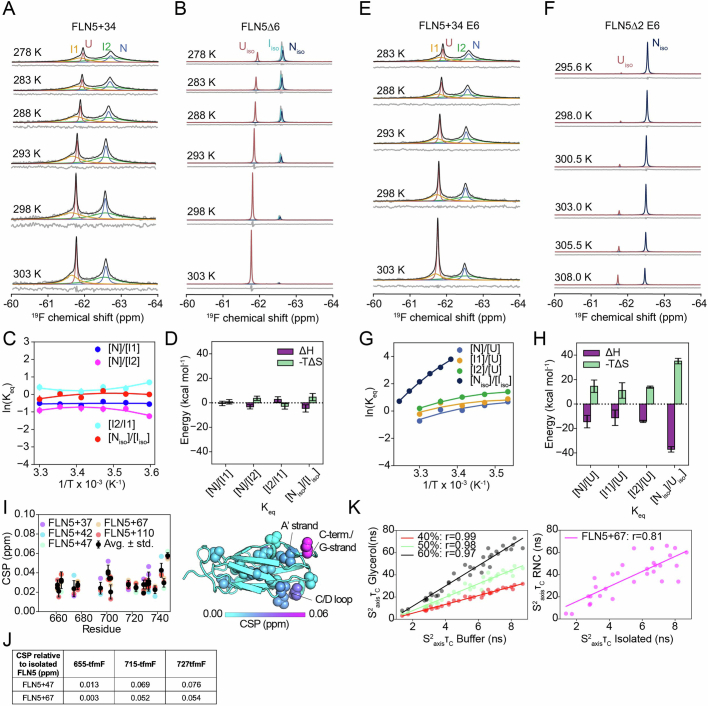
Extended Data Fig. 7. Dependence of the folding equilibrium constant on temperature and structural perturbations observed in the native state on the ribosome.
(A-B) 19F NMR spectra of FLN5 on and off (Δ6 truncation) the ribosome recorded at a 19F-Larmor frequency of 470 MHz. Raw spectra are shown in grey, lineshape fits in colour and the total fit in black. Residuals after fitting are shown below each spectrum. (C-D) Nonlinear fits to a modified Gibbs-Helmholtz equation (see methods) of the equilibrium constants on and off the ribosome measured by 19F NMR (from panels A-B) shown as the mean ± SEM propagated from NMR line shape fits (panel C) and the resulting thermodynamic parameters (mean ± SD from fits, panel D). (E-F) 19F NMR spectra of the FLN5 mutant E6 on and off the ribosome (Δ2 truncation) recorded at a 19F-Larmor frequency of 470 MHz. The FLN5Δ2 E6 was chosen due to its suitable stability in this temperature range to quantify both [U] and [N]. Raw spectra are shown in grey, lineshape fits in colour and the total fit in black. Residues after fitting are shown below each spectrum. (G-H) Nonlinear fits to a modified Gibbs-Helmholtz equation (see methods) of the equilibrium constants on and off the ribosome measured by 19F NMR (from panels E-F) shown as the mean ± SEM propagated from NMR line shape fits (panel G) and the resulting thermodynamic parameters (mean ± SD from fits, panel H). (I) Left: Chemical shift perturbations (CSPs) measured by NMR (1H-13C HMQC) for methyl groups of natively folded FLN5 (RNCs relative to the isolated protein)25. The black datapoints represent the mean ± SD from five different RNC lengths for ease for visualisation. Right: Average CSPs mapped on the crystal structure of FLN593. (J) CSPs (RNC relative to isolated protein) measured for FLN5 labelled with three different 19F-tfmF labelling sites by 19F NMR at linker lengths of 47 and 67 amino acids6. (K) Correlation plots (along with Pearson correlation coefficients) of methyl relaxation parameters (τC) for natively folded FLN525 in different concentrations of glycerol (left panel) and correlating FLN5 on and off the ribosome (right panel).