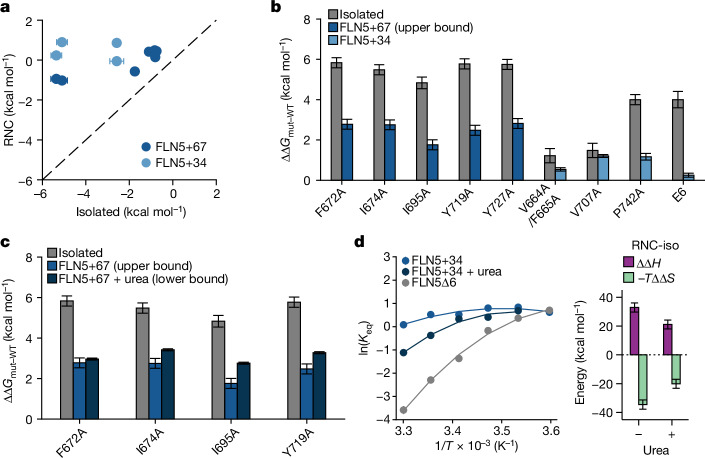
Fig. 5. Destabilizing mutations are buffered by the ribosome.
a, Folding free energies of destabilizing mutants off and on the ribosome (FLN5+34 and FLN5+67 depending on the stability of the mutant) determined by 19F NMR from U and N state populations. b, Destabilization (ΔΔGN-U,mut-WT) of all mutants in isolation compared to the RNC. WT, wild type. c, Destabilization of 4 mutants in isolation, on the ribosome (FLN5+67) and on the ribosome in the presence of 2.5 M urea. d, Left, temperature dependence of the folding equilibrium constant involving the N and U state of isolated FLN5Δ6, FLN5+34 and FLN5+34 in 1.5 M urea measured by 19F NMR fit to a modified Gibbs–Helmholtz equation (Methods). The error bars of individual datapoints (propagated from bootstrapped errors of NMR lineshape analyses) are similar in magnitude to the size of the circles. Right, thermodynamic parameters (T = 298 K) from the nonlinear fits (mean ± s.d.) shown as the difference relative to the isolated protein. Unless stated otherwise, all values in the figure represent the mean ± s.e.m. propagated from NMR lineshape fits.