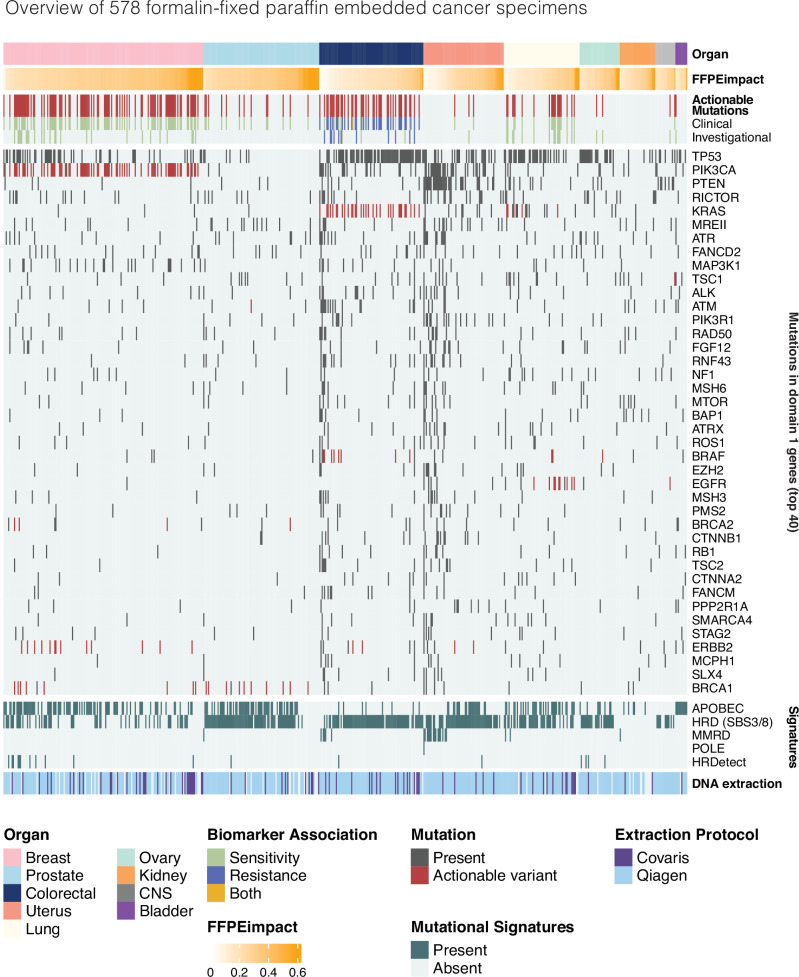
Fig. 4. Overview of Genomics England FFPE cohort.
Samples are arranged by organ type and then by FFPEimpact score (in ascending order). Mutations were considered actionable if they met the criteria discussed in the methodology section. Actionable mutations were divided into Clinical and Investigational depending on ESCAT tier (see the “Methods” section). Mutations in the top 40 mutated Domain 1 genes are presented with actionable variants highlighted. Mutational signatures were grouped by aetiology. APOBEC: APOBEC protein family dysfunction; HRD: homologous recombination deficiency (SBS3 and SBS8); MMRD mismatch repair deficiency, POLE DNA polymerase epsilon dysfunction, HRDetect HRDetect algorithm corrected for artefact as discussed in methodology and a supplemental appendix. DNA extraction protocol compares the two different protocols (Covaris and Qiagen) used for the Genomics England cohort. FFPE formalin-fixed paraffin-embedded.