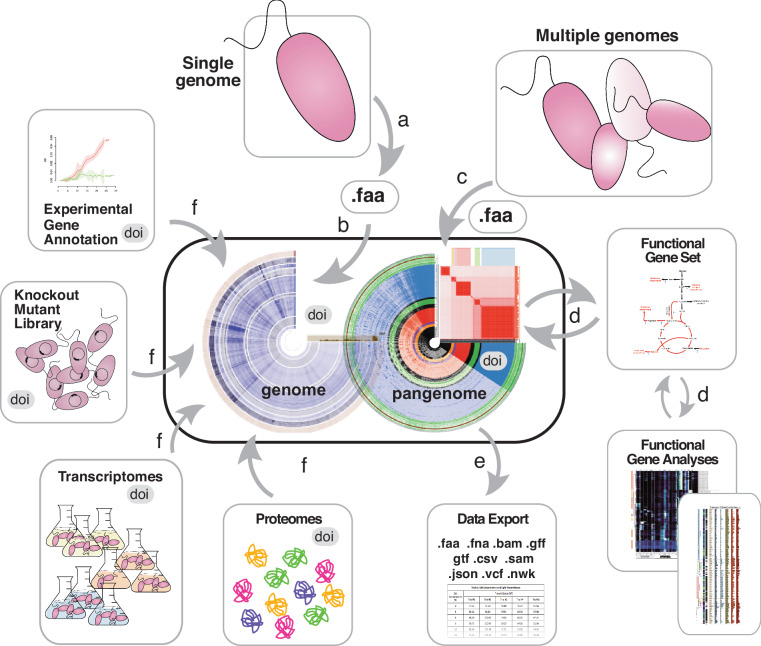
Fig. 1.
Architecture of a Digital Microbe. The genome of a model bacterium is (a) sequenced and (b) assembled and serves as the foundation of a Digital Microbe, a self-contained data package for a collaborative research team or a science community. (c) Alternatively, a pangenomic data package is assembled. (d) Intermediate datasets useful for downstream analyses are stored and reused, and (e) various data files and tables can be exported. (f) The Digital Microbe is iteratively populated with data layers referenced to individual genes, including mapped proteomes, transcriptomes, or gene-specific metadata types such as inventories of mutants or new annotations. Each Digital Microbe can be assigned a DOI (digital object identifier) and be versioned as new gene- or genome-referenced data are added.