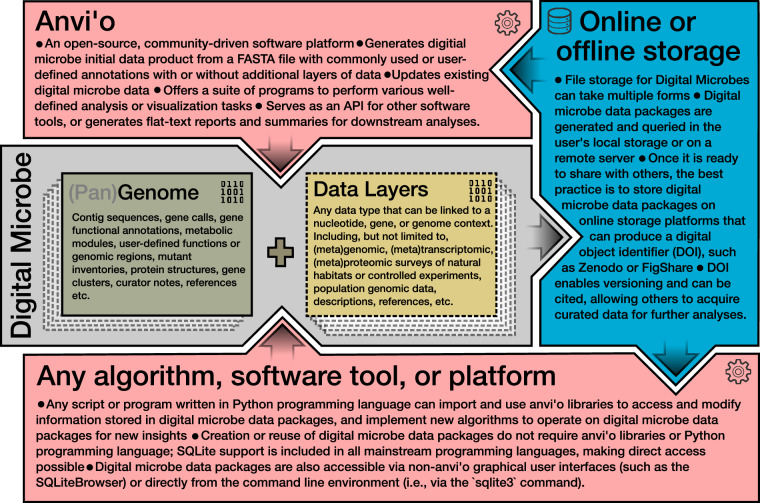
Fig. 2.
Situating the Digital Microbe concept in the existing computational environment. The Digital Microbe approach facilitates collaborative science by: establishing a version-controlled (pan)genomic reference; consolidating and cross-referencing collections of experimental and environmental data associated with a genome or pangenome; facilitating access to reusable intermediate analyses; and providing data export capabilities for transitioning to other programs or analysis software. While each of these features could be established by generating new software, we chose to use the existing open-source software platform anvi’o6, which implements several aspects of a Digital Microbe via (pan)genomic data storage in programmatically-queryable SQLite databases. The concept behind the Digital Microbe framework, however, is independent of any one software platform.