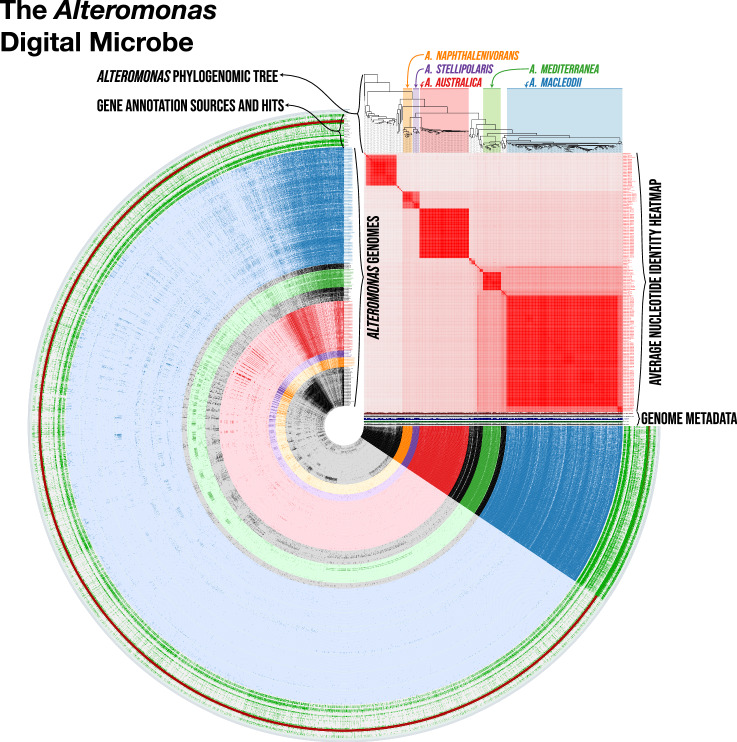
Fig. 5.
The Alteromonas Digital Microbe. Each concentric ring represents one Alteromonas genome, with colored rings identifying genomes from five clades of interest (A. macleodii, A. mediterranea, A. austalica, A. stellioolaris, and A. naphthalenivorans). The outermost green rings depict annotation sources applied to all genomes. Each spoke in the figure represents one gene cluster in the pangenome, with presence/absence denoted by darker/lighter colors, respectively. Genome metadata are shown next to each ring and include total genome length, GC content, completion, number of genes per kbp, and number of gene clusters per genome. The red heatmap above the metadata shows the average nucleotide identity (ANI) percentage scores between genomes. The tree above the ANI heatmap shows the imported phylogenomic tree, with clades of interest color-referenced in the circular portion of the figure. This figure was generated using the anvi’o ‘anvi-display-pan’ from a version of the Alteromonas digital microbe without singleton genes, which is available on Zenodo under 10.5281/zenodo.10421034.