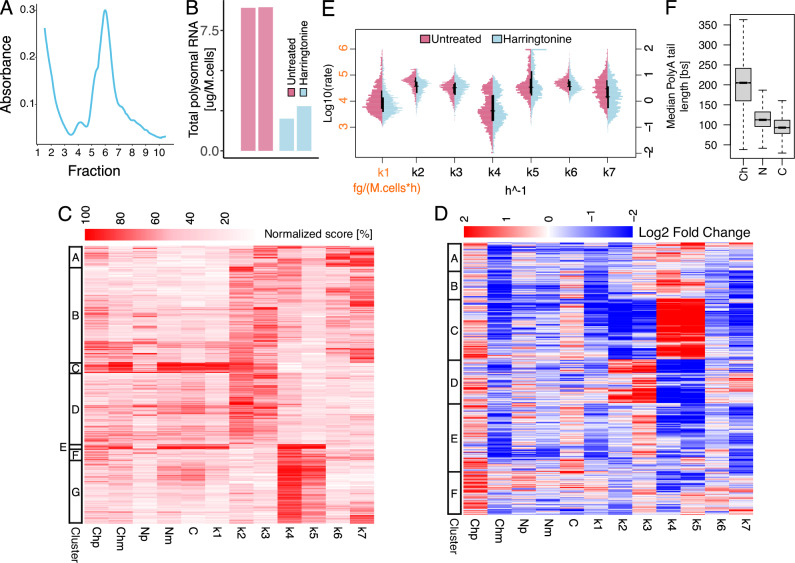
Fig. 5. Blocking the translational machinery hampers RNA export and cytoplasmatic degradation.
A Typical polysome trace for Harringtonine treated cells. B Yield of polysomal RNA in untreated and Harringtonine treated cells; two biological replicates per condition reported. C Heatmap reporting for each of the modelled genes (rows) RNA species abundance and kinetic rates magnitude (columns). For each column, values are saturated between the 1st and 99th percentiles and normalised against the latter. The left bar indicates gene sets similar in abundances and kinetic rates (k-means clustering). D Heatmap depicting genes (rows) abundances and kinetic rates modulations (columns) in response to Harringtonine treatment (log2 fold changes saturated between −2 and +2). The left bar indicates gene sets similar in abundances and kinetic rates modulations (k-means clustering). E Kinetic rates distributions in log10 scale for untreated (purple) and Harringtonine treated (light blue) cells; horizontal and vertical black lines represent medians and interquartile ranges of the distributions, respectively. F Gene-level median polyA tail length distributions across RNA pools. In the figure, Ch, N, and C denote chromatin associated, nucleoplasmic, and cytoplasmic RNA while p and m specify premature and mature forms. Analyses performed in C, E were performed on 1616 genes processed with a model accounting for all the steps of the RNA life cycle except for association with polysomes and polysomal degradation in the Harringtonine treated condition. The analysis reported in D was performed on 896 genes processed with the same model used for C, E in both untreated and Harringtonine treated conditions. The analysis reported in F was performed on genes with at least one profiled polyA tail after replicates pooling (11832, 12242, 11646 genes for Ch, N, and C). For boxplots, the horizontal line represents the median value, the box edges represent the 25th (Q1) and 75th (Q3) percentiles, and the whiskers show the range of data excluding outliers (observations lower that Q1 − 1.5 * interquartile range or larger than Q3 + 1.5 * interquartile range). Source data are provided as a Source Data file.