Figure 3.
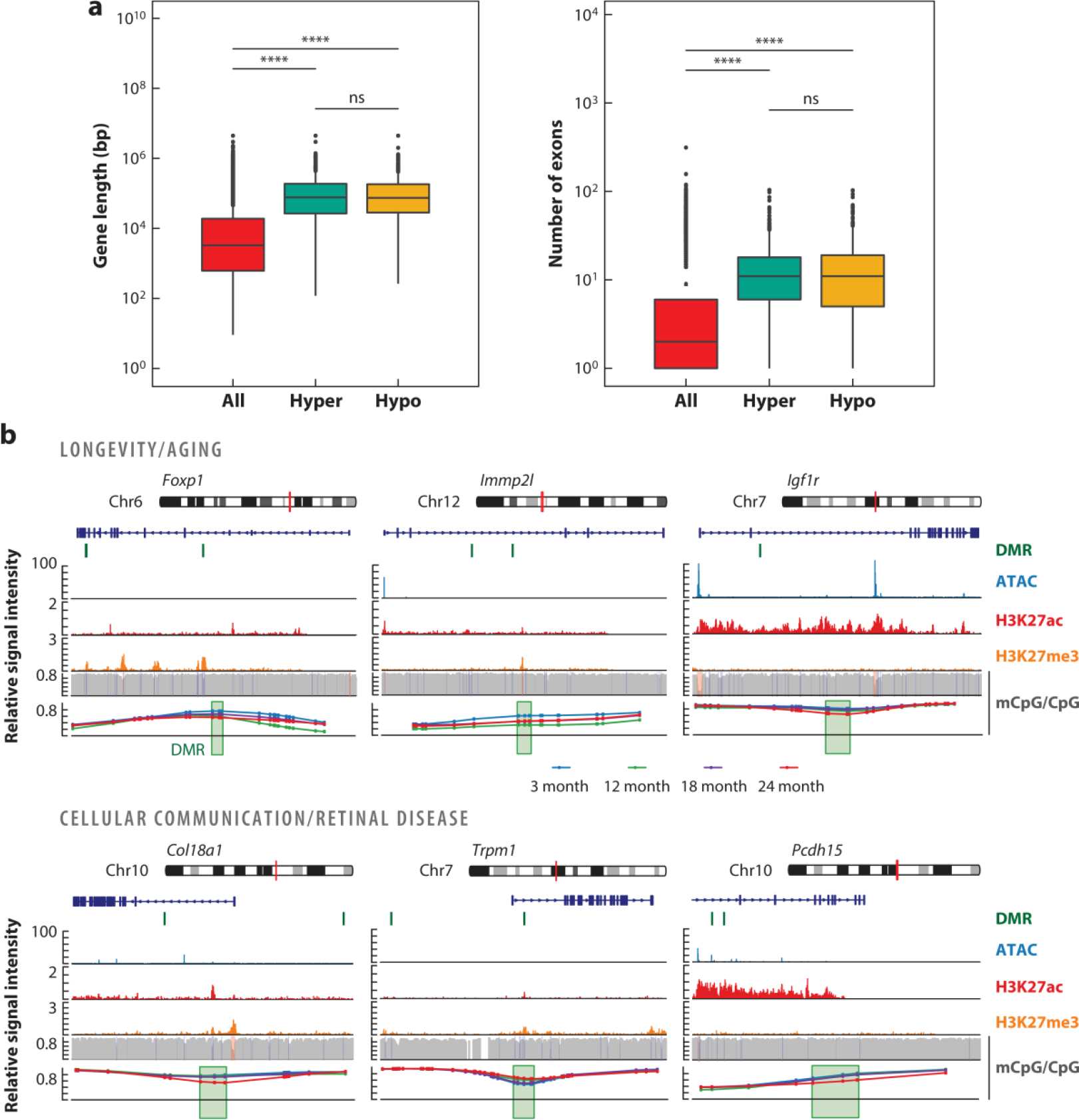
Differential methylation at gene bodies of long genes. Re-analysis of published (Corso-Diaz et al. 2020) differentially methylated regions (DMRs) identified in purified mouse rod photoreceptors at 24 months old compared to at 3 months old is shown. (a) Gene length expressed in base pairs (bp) and exon number of all genes and genes containing DMRs that are hyper- or hypo-methylated with age. ***indicates p < 2 × 10−16; ns indicates not significant. (b) Genome browser view of DMRs at genes involved in aging and longevity (Foxp1, Immp2l, and Igf1r) and cellular communication (Col18a1, Trpm1, and Pcdh15). The red vertical line on the chromosome pictogram indicates the location of the loci presented below. The location of DMRs on or in the vicinity of associated genes is shown by a green vertical line. Signal levels for open chromatin (ATAC-seq), histone modifications associated with active (H3K27ac) and repressed (H3K27me3) regions, and CpG methylation are shown for the three-month time point across the respective genes and their surroundings. The bottom panel represents a zoomed-in view of DMRs (green box) showing methylation levels of each CpG for rods from the 3-month-old (blue), 12-month-old (green), 18-month-old (purple), and 24-month-old (red) retina.
