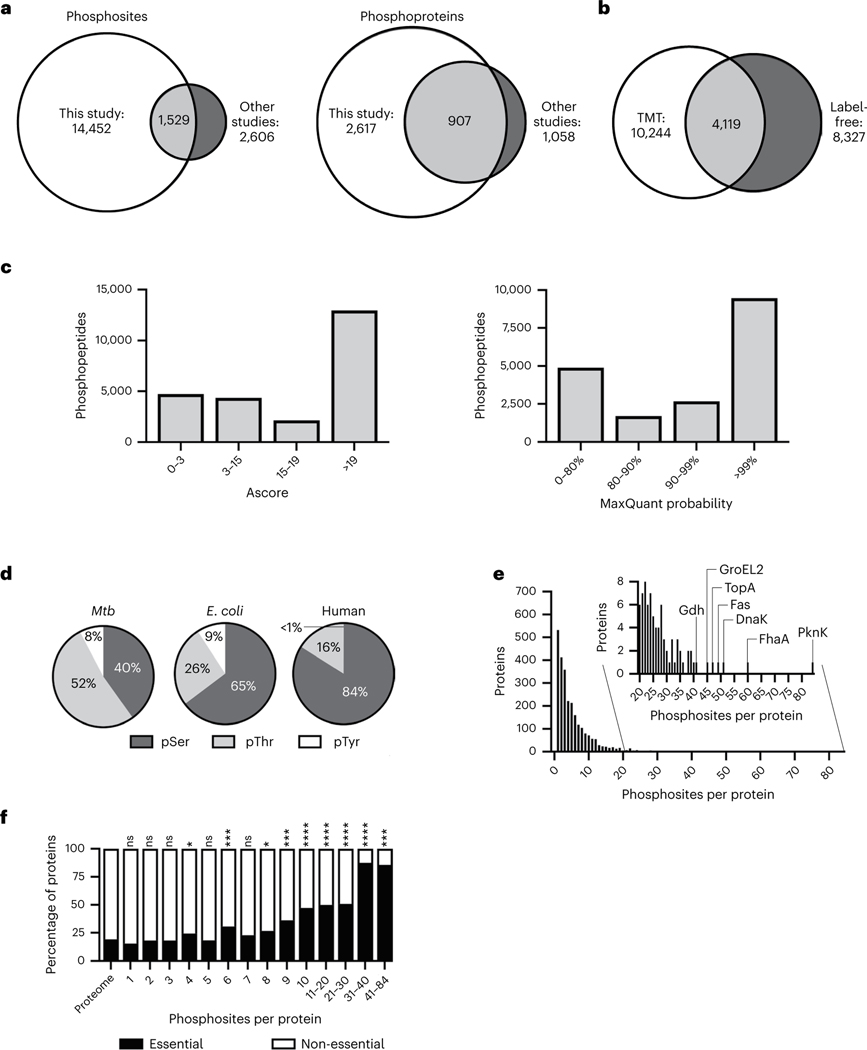
Fig. 1 |. A deep Mtb phosphoproteome.
a, Comparison of the number of unique protein O-phosphorylation sites (left) and proteins (right) identified in this study and previously published. b, Number of unique phosphosites identified by the TMT and label-free approaches. c, Distribution of phosphosite localization Ascore (for the TMT data) and MaxQuant probability (for the label-free data). An Ascore of 19 corresponds to 99% probability of correct localization. d, Relative share of pSer, pThr and pTyr sites in Mtb, E. coli and human cells. e, Number of phosphorylation sites per protein and highly phosphorylated proteins with >20 phosphosites (inset). f, Share of phosphorylated essential gene products stratified by the number of phosphosites. Essentiality was based on transposon mutagenesis (30). P values were calculated using a hypergeometric test for enrichment and are *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.