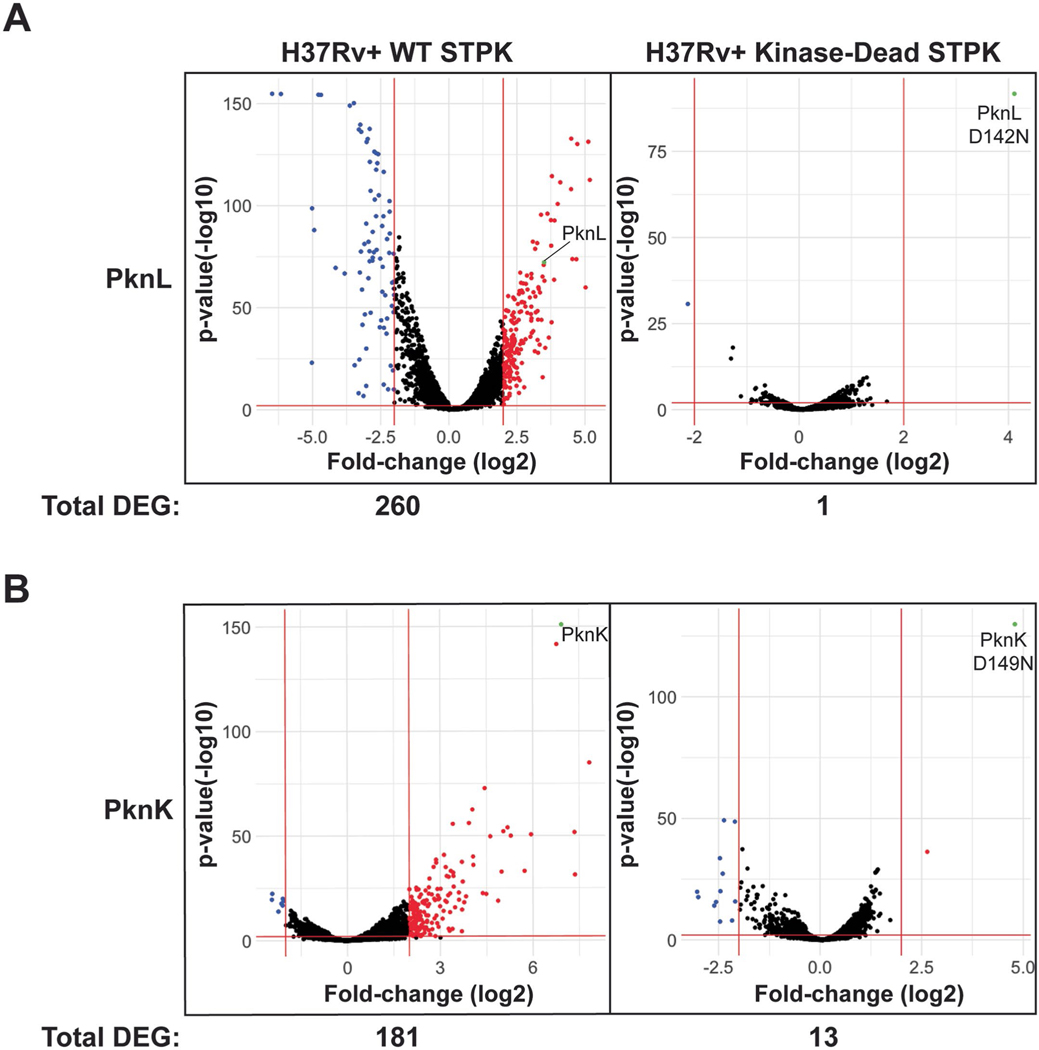
Extended Data Fig. 8 |. Transcriptional effects of a PknL and PknK kinase-dead mutant.
(a) The PknL kinase-dead mutant reduces the DEGs compared to WT OE from 260 to 1. PknL WT and kinase-dead transcripts are highlighted in green, showing induction in both cases. (b) The PknK kinase-dead mutant reduces DEGs compared to WT OE from 181 to 13, suggesting a small transcriptional effect of the PknK malT domain in the OE. PknK WT and kinase-dead transcripts are highlighted in green, showing induction in both cases. Horizontal and vertical red lines show significance cutoffs (>4-fold, P < 0.01) used for further analysis of DEGs. Significance was determined by calculating the geometric mean of five DE tools.