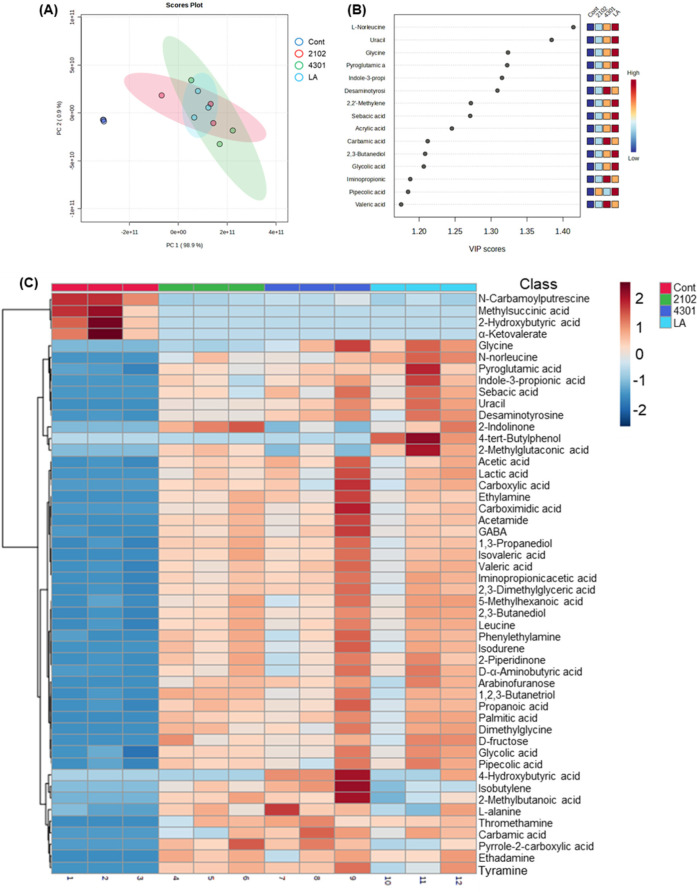
Fig. 3. Comparative analysis of unique metabolite production by probiotics in FIMM.
Following the FIMM cultivation, variations in metabolite profiles across different groups were examined. (A) In the PCA score plots, the analysis revealed that fecal samples from groups subjected to probiotic interventions (LA, 2102, and 4301) clustered together, indicating a shared metabolic response. In contrast, the control group was distinctly clustered, highlighting significant metabolic differentiation from the treated groups. (B) The partial least squares discriminant analysis (PLS-DA) further analyzed these differences, identifying metabolites that drove the separation between treated and untreated groups. Additionally, the colored boxes in (B) and (C) categorized the top 50 abundant metabolites, with varying colors denoting concentration levels, offering an understanding of metabolite fluctuations resulting from FIMM cultivation and probiotic treatments. 2102, Enterococcus faecium IDCC 2102; 4301, Bifidobacterium lactis IDCC 4301; LA, Lactobacillus acidophilus SLAM AK001; PC, principle component; VIP, variable importance plots; GABA, gamma-aminobutyric acid; FIMM, Fermenter for Intestinal Microbiota Model; PCA, principle component analysis.