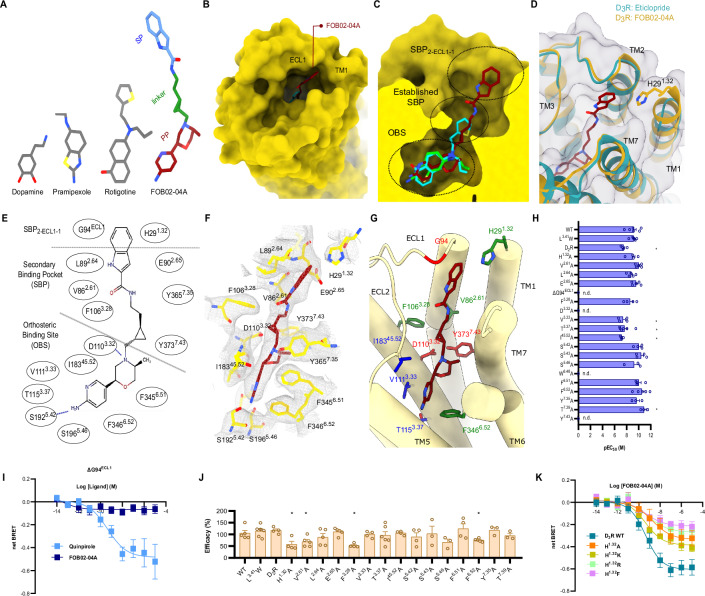
Fig. 3. Binding of the bitopic FOB02-04A to the D3R receptor.
A Schematic of dopamine, pramipexole, rotigotine and the bitopic FOB02-04A ligand shown as sticks and colored by component. B Binding of the secondary pharmacophore (SP) (sticks, dark red) to a groove-shaped pocket at the D3R (yellow, surface representation) formed by ECL1 and TM1. C Two views of a comparison of FOB02-04A (dark red carbon, sticks), pramipexole (green carbon, sticks), and rotigotine (cyan carbon, sticks) binding into the D3R pocket (yellow, surface representation). Dashed circles indicate OBS, established SBP, and the SBP2-ECL1-1 site. D Overall binding mode of the bitopic molecule to the D3R and ordering of TM1 upon bitopic binding. FOB02-04A (dark red, sticks) is displayed on superposed structures of D3R bound to eticlopride (cartoon, cyan) and FOB02-04A (cartoon, yellow) E Schematic of the FOB02-04A binding into the D3R ligand binding pocket. F Binding details of FOB02-04A (dark red, sticks) at the D3R (yellow sticks) with cryo-EM density as gray mesh. G Binding details of FOB02-04A (dark red, sticks) at the D3R (yellow cartoons) with residues at the ligand binding pocket colored by functional effect when mutated to alanine: decreased efficacy – green carbons, decreased potency – blue carbon and non-detectable binding – red carbon. H pEC50 values for alanine mutation of the residues at the ligand binding site in response to GOA activation by FOB02-04A using the TRUPATH assay. All data are means ± SEM of four independent experiments (n = 4) performed in technical triplicates except for D1103.32A, S1965.46A, Y3657.35A, T3697.39A, W3426.48A and Y3737.43A for which there was n = 3, WT, V862.61A, L892.64A, E902.65A, ∆G94ECL1, F3466.52A for there was n = 5, and L1193.41W, T1153.37A for which there was n = 6. *p < 0.05 (one-way ANOVA with Dunnett post hoc analysis) for D2R (p = 0.0081), V1113.33A (p = 0.0049), T1153.37A (p = 0.0074) and I18345.82A (p = 0.0013) and nd - non-detectable. I Concentration-response curve of D3R ΔG94ECL1 upon GOA activation by quinpirole (light blue, n = 4) and FOB02-04A (deep blue, n = 5) (shown as net BRET). All data are means ± SEM of the specified biological replicates, each performed in technical triplicates. J Emax values for alanine mutation of the residues at the ligand binding site in response to GOA activation by FOB02-04A using the TRUPATH assay. Emax values have been normalized to D3R WT. All data are means ± SEM of four independent experiments performed in technical triplicate (n = 4) except for D1103.32A, S1965.46A, Y3657.35A, T3697.39A, W3426.48A, Y3737.43A (n = 3), WT, V862.61A, L892.64A, E902.65A, ∆G94ECL1, F3466.52A (n = 5) and L1193.41W, T1153.37A (n = 6). *p < 0.05 (Holm-Sidak multiple comparisons tests two-tailed p value) for H291.32A (p = 0.016), V862.61A (p = 0.026), F1063.28A (p = 0.003), F3466.52A (p = 0.019). K Concentration-response curves of D3R H291.32A (orange), H291.32F (pink), H291.32K (yellow), and H291.32R (green) upon GOA activation by FOB02-04A (shown as net BRET). All data are means ± SEM derived from three independent experiments (n = 3), each performed in technical triplicate except for H291.32A (n = 4). All source data within this figure is provided as a Source Data file.