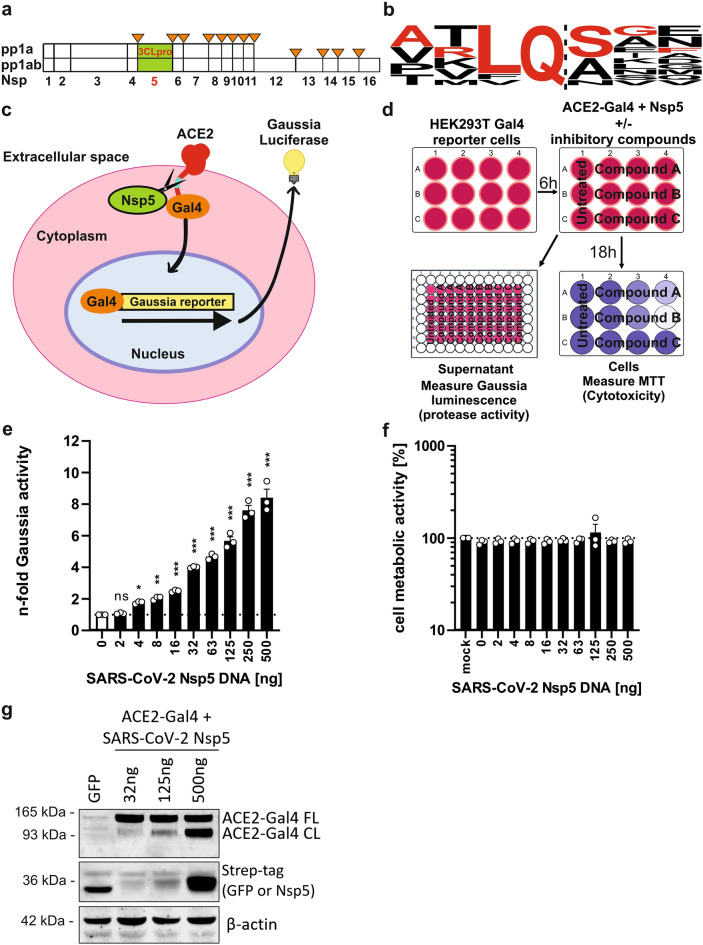
Fig. 1.
Design and performance of the Nsp5 reporter assay. (a) Location of Nsp5 cleavage sites in coronavirus polyprotein 1ab. (b) Sequence logo of the Nsp5 recognition site based on SARS-CoV-2 1ab polyprotein. The sequence of representative cleavage site selected for testing is indicated in red. (c) Principle of the reporter assay. Human ACE2 fused to transcription factor Gal4 is expressed in HEK293T cells containing a stable Gaussia luciferase reporter downstream of 5 × Gal4 binding sites. The fusion protein contains Nsp5 recognition site in the flexible glycine linker region. Upon cleavage, Gal4 fused to amino acids 364–550 of mouse NF-kB translocates into the nucleus where it activates the transcription of Gaussia luciferase, which is subsequently secreted into cell supernatant. (d) Experimental outline of the Nsp5 reporter assay. HEK293T cells containing the stable Gaussia luciferase reporter downstream of the 5 × Gal4 binding sites are co-transfected with construct encoding ACE2-Gal4 fusion protein, and varying amounts of Strep-tagged Nsp5 or GFP (negative control). At 18-20 h post transfection supernatants are harvested for luminescence measurement of Gaussia luciferase activity (relative light units per second; rlu/s) using coelenterazine substrate, while the cells are subjected to 2,5-diphenyl-2H-tetrazolium bromide (MTT) assay measuring cell metabolic activity. (e) Normalized n-fold increase in Gaussia luciferase activity over background (GFP + reporter only) by transfected SARS-CoV-2 Nsp5, and (f) corresponding MTT assay results measured at 18 h post transfection. (g) Western blot of transfected cell lysates showing dose-dependent ACE2-Gal4 reporter cleavage (FL full length, CL cleaved protein detected by ACE2 staining). Lane 1 contained GFP only control. Mean of 3 independent experiments + SD. *P < 0.05; **P < 0.01; ***P < 0.001, ns, P > 0.05. Unpaired Student’s t-test.