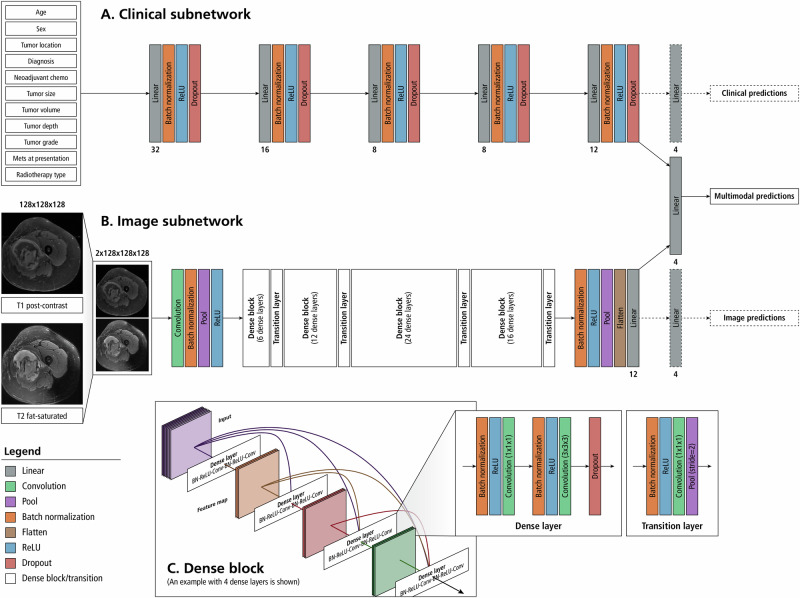
Fig. 4. Architecture of our multimodal neural network model.
A deep neural network (A) will interpret the 11 clinical variables and a 2-channel convolutional neural network (DenseNet-121) analyzes the MRI input (B). Image features from T1 and T2 MRI sequences are extracted by the convolutional neural network and this information is concatenated along with the features extracted from the clinical variables. Analysis of the combined feature set is used to predict the risk of distant metastases and overall survival. Gradient blending is used to moderate the weight updates between modalities. Dashed lines are used to indicate connections that are only present during training to facilitate Gradient Blending. 1A: Clinical Subnetwork Model. A deep neural network is implemented to extract features from a vector of clinical variables corresponding to the patient. Numbers under the linear layers correspond to the number of output features for those linear layers. The clinical model extracts 12 features that will be used for the multimodal prediction. 1B: Image Subnetwork Model. T1 post contrast and T2 fat-sat MRI sequences are concatenated along the channel dimension prior to being fed through a 2-channel DenseNet-121 model. Twelve features are extracted for use in the multimodal prediction. The numbers in each dense block correspond to the number of dense layers within that dense block. The architecture presented is representative of a 3-dimensional, 2-channel densenet-121 with 12 output neurons. Because the model is being used as a feature extractor rather than a classifier, the size of the output layer is a tunable parameter and not limited to the number of predictions made by the multimodal output head. 1C: Dense Block– Dense blocks consists of a series of dense layers. Within each dense block, the resolution of the feature map is constant. This allows all dense layers within a dense block to contain feed-forward bypass connections to every other dense layer in that dense block. These features are concatenated at the input of each dense layer. Transition layers are placed between dense blocks. Transition layers use 1x1x1 convolutions to act as channel pooling layers, reducing the number of feature maps by a factor of 2. In addition, stride 2 average pooling layers are used which reduce the resolution in all spatial dimensions by a factor of 2.