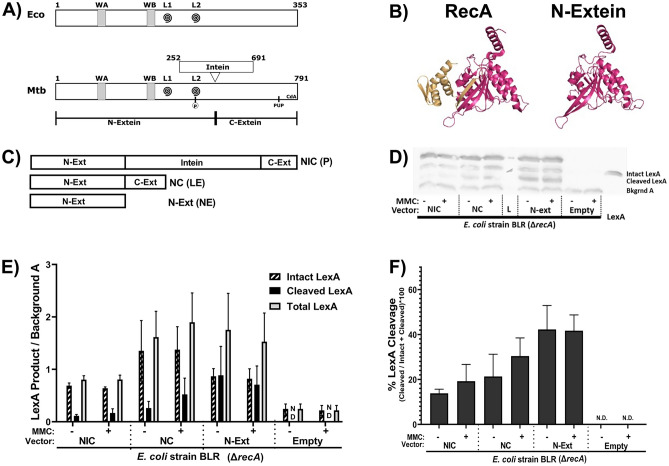
Fig. 5.
The N-extein product can activate the SOS response in E. coli. (A) Domain architecture map of (top) E. coli RecA protein and (bottom) Mtb RecA protein. WA and WB indicate the Walker A and B motifs. L1 and L2 (swirls) indicate loop 1 and loop 2. The residue in Mtb RecA that can be phosphorylated is in L2. The pupylation site is marked as pup, and the cyclic-di-AMP binding area is labeled CdA. (B) (left) Previously crystallized Mtb RecA structure from Datta et al. (PDB: 1mo3). The N-extein is colored red, and the C-extein is shaded gold. (right) I-Tasser derived model of folded N-extein colored in red. (C–F) Production of LexA and LexA cleavage in E. coli ∆recA BLR strains complemented with Mtb RecA constructs driven by the native Mtb promoters: NIC—recA including intein; NC—inteinless recA; N-Ext—N-extein producing only. (C) Schematic of the three constructs. (D) Representative western blot probing for LexA. (E) Densitometric analysis of LexA western blots normalized to an unchanging background band (Bkgrnd A). (F) Quantified cleavage of LexA from western blot analysis. N.D. – Not detected. Error bars indicate SEM of 3 biological replicates.