Figure 2.
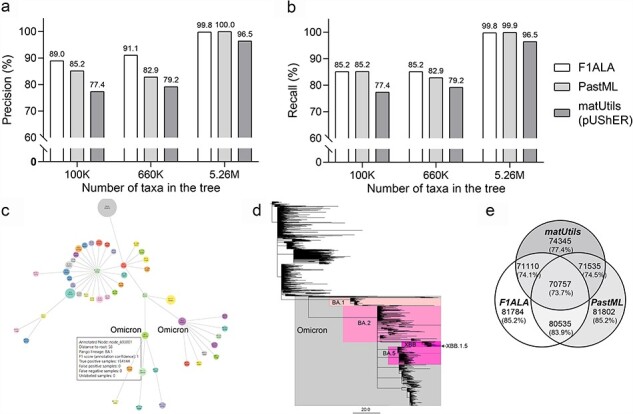
Accuracy of ALA for the 100K-, 660K-, and 5.26M-taxa SARS-CoV-2 phylogenies. (a) Precision (b) Recall for ALA by F1ALA, PastML, and matUtils (pUShER). (c) The ALA using the 5.26M-taxa SARS-CoV-2 phylogeny by F1ALA, showing the top 50 lineages by the number of assigned taxa, with the Omicron lineage highlighted (branch lengths are not to scale to allow differentiation of the lineages). Annotation information (annotation node, distance to tree root, F1-score, and number of TPs) is shown when a mouse hovers over the nodes displayed in a browser. (d) The collapsed tree of 2277 PANGO lineages from the 5.26M-taxa SARS-CoV-2 phylogeny. Each lineage is represented by its annotation node in the tree. Branch length shows the number of mutations (instead of substitution rate) (McBroome et al. 2021) and Omicron sublineages (BA.1, BA.2, BA.5, and XBB.1.5) are highlighted. (e) Venn diagram showing the number of individual and shared TPs (proportions over all taxa) for the annotations by F1ALA, PastML, and matUtils.
